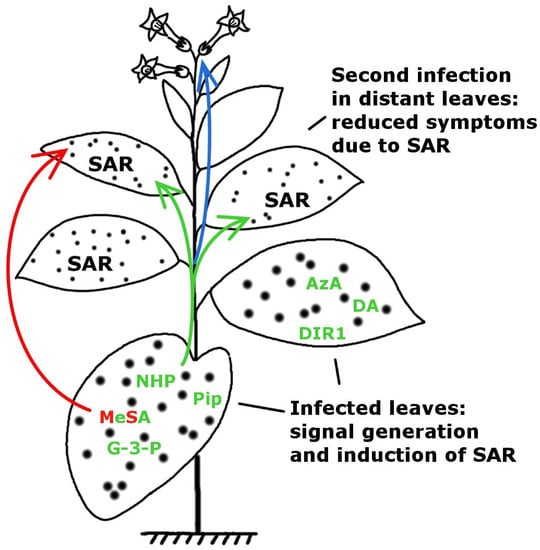
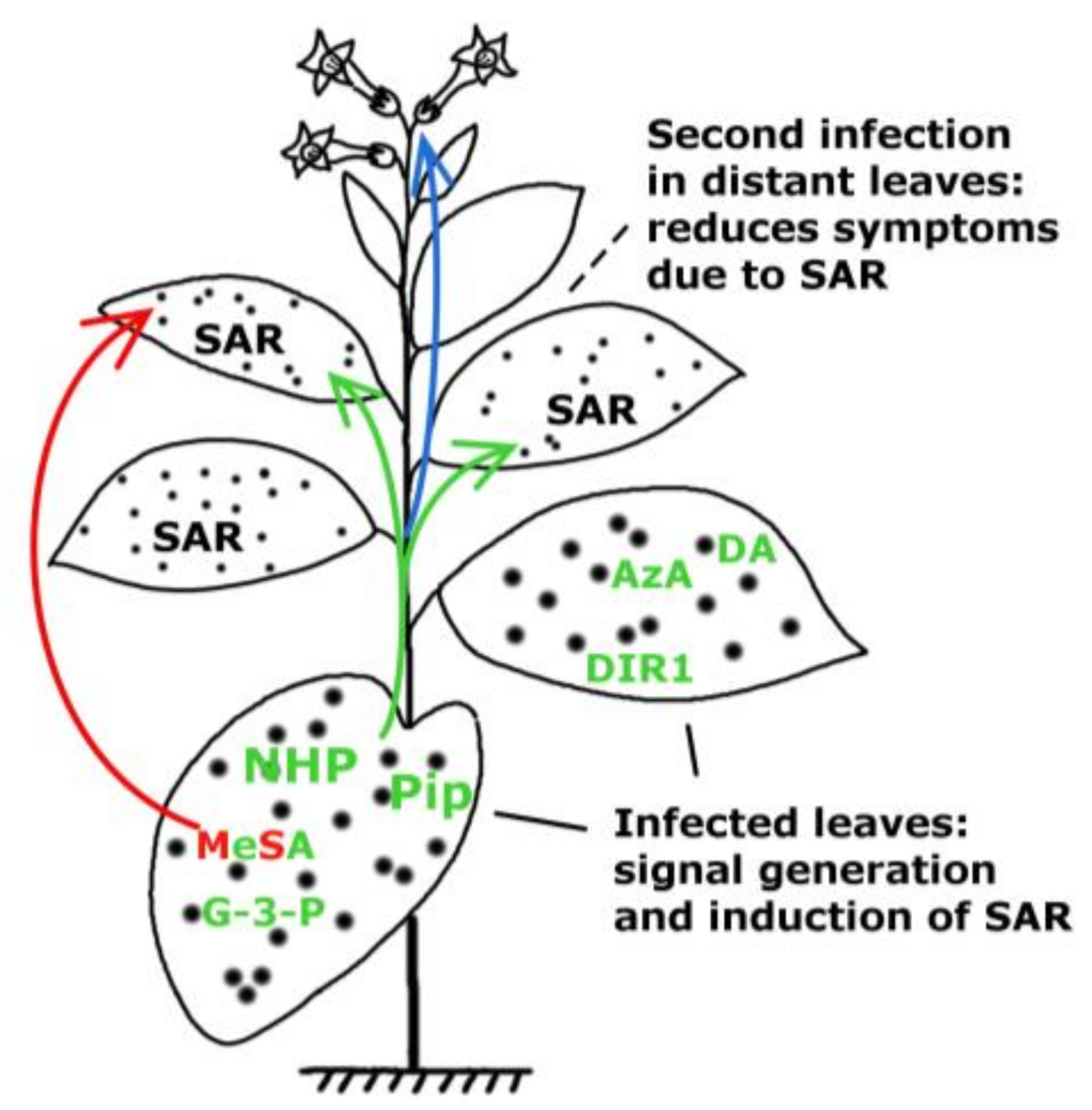
Signals of Systemic Immunity in Plants: Progress and Open Questions
Abstract
:1. Introduction
2. Multiple Signalling by Chemically Diverse Compounds during SAR Induction
2.1. DIR1 (DEFECTIVE IN INDUCED RESISTANCE1)
2.2. Salicylic Acid (SA) and Methyl Salicylate (MeSA)
2.3. Lipid-Derived Signalling
2.3.1. Glycerol-3-Phosphate Dependent Factor (G3P-Dependent Factor)
2.3.2. Azelaic Acid (AzA)
2.4. Dehydroabietinal (DA)
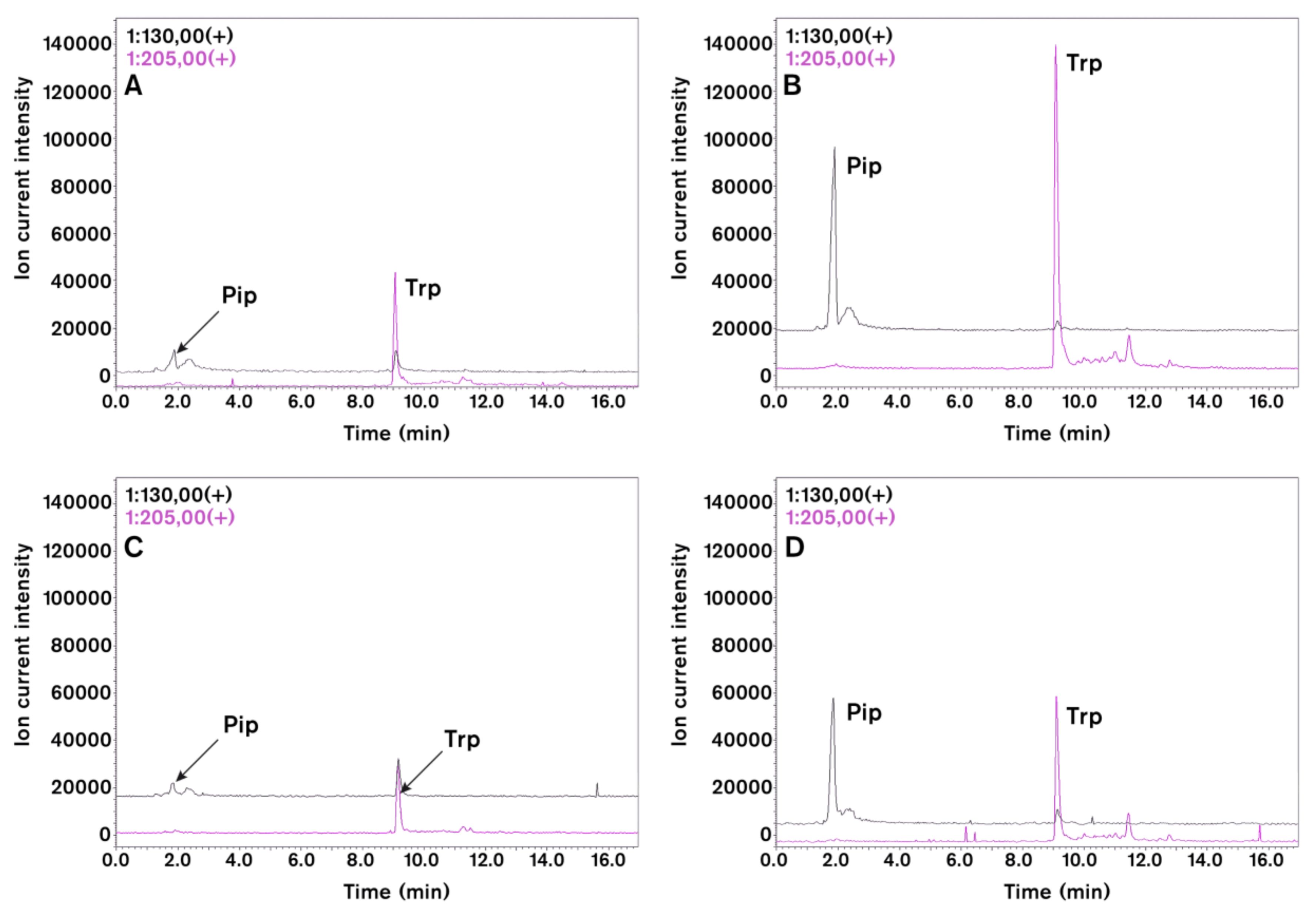
2.5. Pipecolic Acid (Pip) and N-hydroxypipecolic Acid (NHS)
3. Role of Light in SAR Induction: Light Intensity, Timing of Exposition and Spectral Distribution
4. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Winter, P.S.; Bowman, C.E.; Villani, P.J.; Dolan, T.E.; Hauck, N.R. Systemic acquired resistance in moss: Further evidence for conserved defence mechanisms in plants. PLoS ONE 2014, 9, e101880. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.F. Systemic acquired resistance induced by localized virus infections in plants. Virology 1961, 14, 340–358. [Google Scholar] [CrossRef]
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, A.L.; Stassen, J.H.; Furci, L.; Smith, L.M.; Ton, J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016, 88, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Conrath, U. Molecular aspects of defence priming. Trends Plant Sci. 2011, 16, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Dong, X. How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 2012, 12, 89–100. [Google Scholar] [CrossRef] [PubMed]
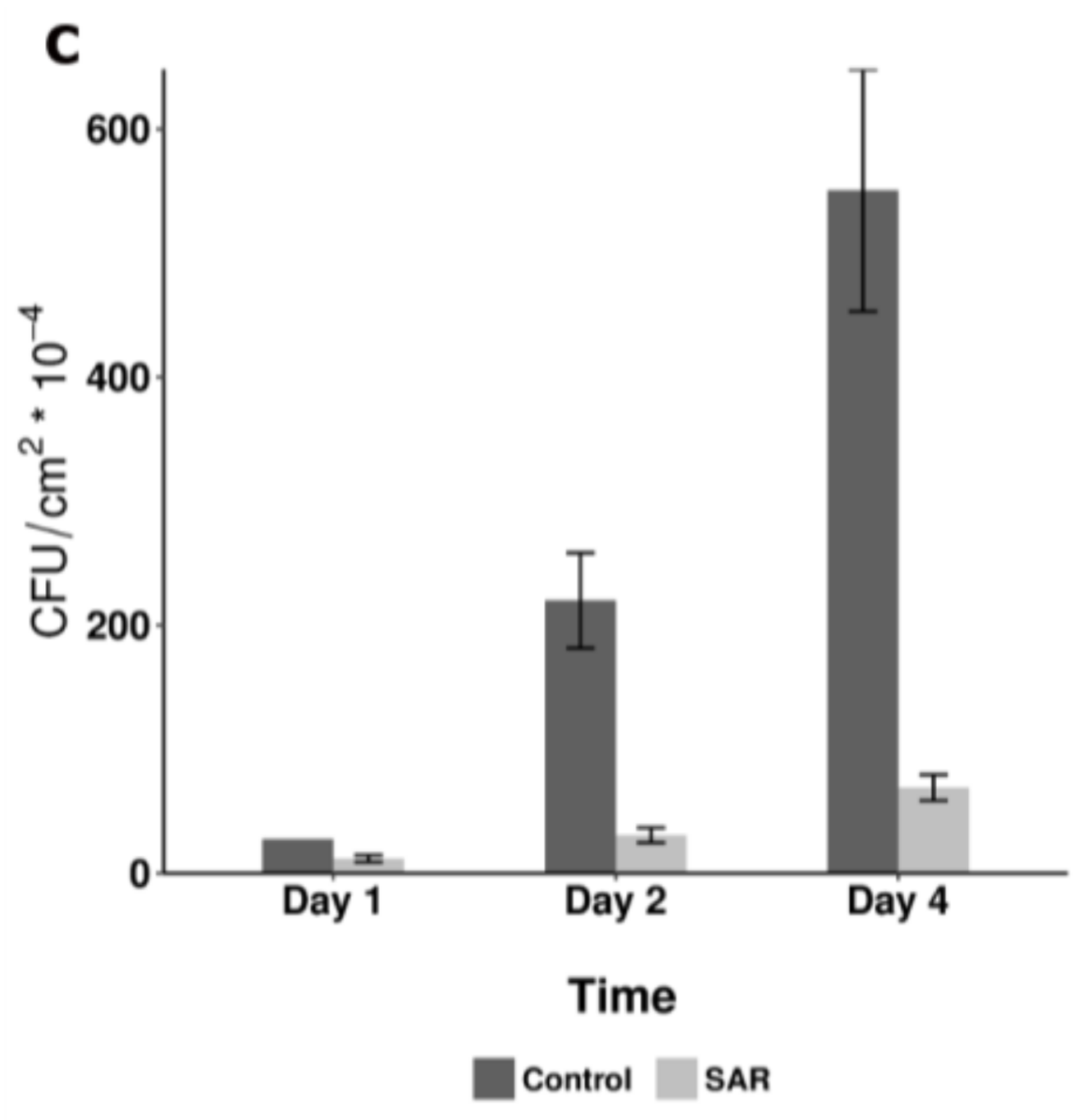
- Shah, J.; Zeier, J. Long-distance communication and signal amplification in systemic acquired resistance. Front. Plant Sci. 2013, 4, 30. [Google Scholar] [CrossRef] [PubMed]
- Jaskiewicz, M.; Conrath, U.; Peterhänsel, C. Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 2011, 12, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.K.; Paiva, N.T.; Lamb, C.J.; Dixon, R.A. Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol. Mol. Plant Pathol. 1999, 55, 121–130. [Google Scholar] [CrossRef]
- Shulaev, V.; Silverman, P.; Raskin, I. Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 1997, 385, 718–721. [Google Scholar] [CrossRef]
- Park, S.W.; Kaimoyo, E.; Kumar, D.; Mosher, S.; Klessig, D.F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 2007, 318, 113–116. [Google Scholar] [CrossRef] [PubMed]
- Manosalva, P.M.; Park, S.W.; Forouhar, F.; Tong, L.; Fry, W.E.; Klessig, D.F. Methyl esterase 1 (stmes1) is required for systemic acquired resistance in potato. Mol. Plant Microbe Interact. 2010, 23, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, A.M.; Doerner, P.; Dixon, R.A.; Lamb, C.J.; Cameron, R.K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 2002, 419, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Isaacs, M.; Cameron, R.K. Plasmodesmata-located protein overexpression negatively impacts the manifestation of systemic acquired resistance and the long-distance movement of Defective in Induced Resistance1 in Arabidopsis. Plant Biol. 2015, 17, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.; Venables, B.; Petros, R.A.; Nalam, V.; Li, M.; Wang, X.; Takemoto, L.J.; Shah, J. An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 2012, 71, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.W.; Tschaplinski, T.J.; Wang, L.; Glazebrook, J.; Greenberg, J.T. Priming in systemic plant immunity. Science 2009, 324, 89–91. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Soares, J.M.; Mandal, M.K.; Wang, C.; Chanda, B.; Gifford, A.N.; Fowler, J.S.; Navarre, D.; Kachroo, A.; Kachroo, P. A feedback regulatory loop between G3P and lipid transfer proteins DIR1 and AZI1 mediates azelaic-acid-induced systemic immunity. Cell Rep. 2013, 3, 1266–1278. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Welti, R.; Shah, J. The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 2004, 16, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Chanda, B.; Xia, Y.; Mandal, M.K.; Yu, K.; Sekine, K.T.; Gao, Q.M.; Selote, D.; Hu, Y.; Stromberg, A.; Navarre, D. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 2011, 43, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Návarová, H.; Bernsdorff, F.; Doring, A.C.; Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 2012, 24, 5123–5141. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Kim, D.; Bernsdorff, F.; Ajami-Rashidi, Z.; Scholten, N.; Schreiber, S.; Zeier, T.; Schuck, S.; Reichel-Deland, V.; Zeier, J. Biochemical principles and functional aspects of pipecolic acid biosynthesis in plant immunity. Plant Physiol. 2017, 174, 124–153. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, F.; Doring, A.C.; Gruner, K.; Schuck, S.; Brautigam, A.; Zeier, J. Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Song, J.T.; Lu, H.; McDowell, J.M.; Greenberg, J.T. A key role for ALD1 in activation of local and systemic defenses in Arabidopsis. Plant J. 2004, 40, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jordá, L.; Maldonado-Alconada, A.M.; Satioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contracting roles of the apoplastic ASPARTYL PROTEASE APOPLASTIC, ENHANCED DISEASE SUSCEPTIBILITY1-DEPENDENT and LEGUME LECTIN-LIKE PROTEIN in Arabidopsis systemic acquired resistance. Plant Physiol. 2014, 165, 791–809. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Park, Y.-J.; Seo, P.J.; Kim, J.-H.; Sim, H.-J.; Kim, S.-K.; Park, C.-M. Systemic immunity requires SnRK2.8-mediated nuclear import of NPR1 in Arabidopsis. Plant Cell 2015, 37, 3425–3438. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ádám, A.L.; Nagy, Z.Á. A szisztemikus szerzett rezisztencia szignálátvitele: Eredmények és kihívások. Signal transduction of systemic acquired resistance: Results and new challenges. Növényvédelem Plant Prot. 2016, 77, 435–461. [Google Scholar]
- Jenns, A.; Kuć, J. Graft transmission of systemic resistance of cucumber to anthracnose induced by Colletotrichum lagenarium and tobacco necrosis virus. Phytopathology 1979, 69, 753–756. [Google Scholar] [CrossRef]
- Nagy, Z.Á.; Kátay, G.; Gullner, G.; Ádám, A.L. Evaluation of TMV lesion formation and timing of signal transduction during induction of systemic acquired resistance (SAR) in tobacco with a computer-assisted method. In Biotic and Abiotic Stress—Recent Advances and Future Perspectives; Shanker, A.K., Shanker, C., Eds.; InTech: London, UK, 2016; pp. 363–372. [Google Scholar]
- Nagy, Z.Á.; Kátay, G.; Gullner, G.; Király, L.; Ádám, A.L. Azelaic acid accumulates in phloem exudates of TMV-infected tobacco leaves, but its application does not induce local or systemic resistance against selected viral and bacterial pathogens. Acta Physiol. Plant. 2017, 39, 9. [Google Scholar] [CrossRef]
- Nagy, Z.Á.; Jung, A.; Varga, Z.; Kátay, Gy.; Ádám, A. Effect of artificial light conditions on local and systemic resistance response of tobacco to TMV infection. Not. Bot. Horti. Agrobot. Cluj-Napoca 2017, 45, 270–275. [Google Scholar] [CrossRef]
- Choi, J.; Tanaka, K.; Cao, Y.; Qi, Y.; Qiu, J.; Liang, Y.; Lee, S.Y.; Stacey, G. Identification of a plant receptor for extracellular ATP. Science 2014, 343, 290–294. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Choi, J.; Cao, Y.; Stacey, G. Extracellular ATP acts as a damage-associated molecular pattern (DAMP) signal in plants. Front. Plant Sci. 2014, 5, 446. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Klauser, D.; Niehl, A.; Domínguez-Ferreras, A.; Chinchilla, D.; Boller, T.; Heinlein, M.; Hann, D.R. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 2013, 26, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Aan den Toorn, M.; Albrecht, C.; de Vries, S. On the origin of SERKs: Bioinformatics analysis of the somatic embryogenesis receptor kinases. Mol. Plant 2015, 8, 762–782. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Gou, X.; Yuan, T.; Lin, H.; Asami, T.; Yoshida, S.; Russell, S.D.; Li, J. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 2007, 17, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Roux, M.; Schwessinger, B.; Albrecht, C.; Chinchilla, D.; Jones, A.; Holton, N.; Malinovsky, F.G.; Tör, M.; de Vries, S.; Zipfel, C. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell 2011, 23, 2440–2455. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis. Plant J. 2007, 50, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Kátay, Gy.; Mergenthaler, E.; Viczián, O.; Nagy, Z.Á.; Ádám, A.L. Centre for Agricultural Research, Hungarian Academy of Sciences, Plant Protection Institute, Budapest, Hungary. Different aspects of systemic immunity in tobacco. Unpublished work. 2018. [Google Scholar]
- Divéki, Z.; Salánki, K.; Balázs, E. The necrotic pathotype of the Cucumber mosaic virus (CMV) ns strain is solely determined by amino acid 461 of the 1a protein. Mol. Plant Microbe Interact. 2004, 17, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Vogel-Adghough, D.; Stahl, E.; Navarova, H.; Zeier, J. Pipecolic acid enhances resistance to bacterial infection and primes salicylic acid and nicotine accumulation in tobacco. Plant Signal. Behav. 2013, 8, e26366. [Google Scholar] [CrossRef] [PubMed]
- Kovalev, N.; Pogany, J.; Nagy, P.D. Template role of double-stranded RNA in tombusvirus replication. J. Virol. 2014, 88, 5638–5651. [Google Scholar] [CrossRef] [PubMed]
- Son, K.N.; Liang, Z.; Lipton, H.L. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-stranded RNA viruses. J. Virol. 2015, 89, 9383–9392. [Google Scholar] [CrossRef] [PubMed]
- Niehl, A.; Wyrsch, I.; Boller, T.; Heinlein, M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 2016, 211, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Ziebell, H.; Carr, J.P. Effects of dicer-like endoribonucleases 2 and 4 on infection of Arabidopsis thaliana by Cucumber mosaic virus and a mutant virus lacking the 2b counter-defence protein gene. J. Gen. Virol. 2009, 90, 2288–2292. [Google Scholar] [CrossRef] [PubMed]
- Henderson, I.R.; Zhang, X.; Lu, C.; Johnson, L.; Meyers, B.C.; Green, P.J.; Jacobsen, S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006, 38, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Park, Y.S.; Lee, S.; Song, G.C.; Ryu, C.M. Bacterial RNAs activate innate immunity in Arabidopsis. New Phytol. 2015, 209, 785–797. [Google Scholar] [CrossRef] [PubMed]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of PATHOGENESIS-RELATED PROTEIN1. Trends Plant Sci. 2017, 10, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Gamir, J.; Darwiche, R.; Van’t Hof, P.; Choudhary, V.; Stumpe, M.; Schneiter, R.; Mauch, F. The sterol-binding activity of PATHOGENESIS-RELATED PROTEIN1 reveals the mode of action of an antimicrobial protein. Plant J. 2016, 89, 502–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Lee, C.Y.; Cheng, K.T.; Chang, W.H.; Huang, R.N.; Nam, H.G.; Chen, Y.R. Quantitative peptidomics study reveals that a wound-induced peptide from PR-1 regulates immune signalling in tomato. Plant Cell 2014, 26, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Klessig, D.F.; Tian, M.; Choi, H.W. Multiple targets of salicylic acid and its derivatives in plants and animals. Front. Immunol. 2016, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Champigny, M.J.; Isaacs, M.; Carella, P.; Faubert, J.; Fobert, P.; Cameron, R.K. Long distance movement of DIR1 and investigation of the role of DIR1-like during systemic acquired resistance in Arabidopsis. Front. Plant Sci. 2013, 4, 230. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Kempthorne, C.J.; Wilson, D.C.; Isaacs, M.; Cameron, R.K. Exploring the role of DIR1, DIR1-like and other lipid transfer proteins during systemic immunity in Arabidopsis. Physiol. Mol. Plant Pathol. 2017, 9, 49–57. [Google Scholar] [CrossRef]
- Cecchini, N.M.; Steffes, K.; Schlappi, M.R.; Gifford, A.N.; Greenberg, J.T. Arabidopsis AZI1 family proteins mediate signal mobilization for systemic defence priming. Nat. Commun. 2015, 6, 7658. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, M.; Carella, P.; Faubert, J.; Rose, J.K.C.; Cameron, R.K. Orthology analysis and in vivo complementation studies to elucidate the role of DIR1 during systemic acquired resistance in Arabidopsis thaliana and Cucumis sativus. Front. Plant Sci. 2016, 7, 566. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; von Dahl, C.C.; Park, S.W.; Klessig, D.F. Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and tobacco. Plant Physiol. 2011, 155, 1762–1768. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Yu, K.; Gao, Q.-M.; Wilson, R.V.; Navarre, D.; Kachroo, P.; Kachroo, A. Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 2012, 3, 224. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Merl-Pham, J.; Wilson, D.C.; Dey, S.; Hauck, S.M.; Vlot, A.C.; Cameron, R.K. Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiol. 2016, 171, 1495–1510. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of salicylic acid for the induction of systemic acquired resistance. Science 1993, 261, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef] [PubMed]
- Métraux, J.P.; Signer, H.; Ryals, J.; Ward, E.; Wyss-Benz, M.; Gaudin, J.; Raschdorf, K.; Schmid, E.; Blum, W.; Inverardi, B. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 1990, 250, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Delaney, T.P.; Uknes, S.; Vernooij, B.; Friedrich, L.; Weymann, K.; Negrotto, D.; Gaffney, T.; Gut-Rella, M.; Kessmann, H.; Ward, E.; et al. A central role of salicylic acid in plant disease resistance. Science 1994, 266, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, S.; Ding, P.; Wang, D.; Cheng, Y.T.; He, J.; Gao, M.; Xu, F.; Li, Y.; Zhu, Z.; et al. Control of salicylic acid synthesis and systemic acquired resistance by two members of a plant-specific family of transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18220–18225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tsuda, K.; Truman, W.; Sato, M.; Nguyen le, V.; Katagiri, F.; Glazebrook, J. CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 2011, 67, 1029–1041. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Métraux, J.P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 1999, 11, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565. [Google Scholar] [CrossRef] [PubMed]
- Nawrath, C.; Heck, S.; Parinthawong, N.; Métraux, J.P. EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 2002, 14, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Koo, Y.J.; Kim, M.A.; Kim, E.H.; Song, J.T.; Jung, C.; Moon, J.K.; Kim, J.H.; Seo, H.S.; Song, S.I.; Kim, J.K.; et al. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol. Biol. 2007, 64, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Meuwly, P.; Molders, W.; Buchala, A.; Metraux, J.P. Local and systemic biosynthesis of salicylic acid in infected cucumber plants. Plant Physiol. 1995, 109, 1107–1114. [Google Scholar] [CrossRef] [PubMed]
- Pallas, J.A.; Paiva, N.L.; Lamb, C.; Dixon, R.A. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 1996, 10, 281–293. [Google Scholar] [CrossRef]
- Catinot, J.; Buchala, A.; Abou-Mansour, E.; Metraux, J.P. Salicylic acid production in response to biotic and abiotic stress depends on isochorismate in Nicotiana benthamiana. FEBS Lett. 2008, 582, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.B.; Hammerschmidt, R.; Zook, M.N. Systemic induction of salicylic acid accumulation in cucumber after inoculation with Pseudomonas syringae pv. syringae. Plant Physiol. 1991, 97, 1342–1347. [Google Scholar] [CrossRef] [PubMed]
- Vernooij, B.; Friedrich, L.; Morse, A.; Reist, R.; Kolditz-Jawhar, R.; Ward, E.; Uknes, S.; Kessmann, H.; Ryals, J. Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance. Plant Cell 1994, 6, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Attaran, E.; Zeier, T.E.; Griebel, T.; Zeier, J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 2009, 21, 954–971. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Roy, S.; Giri, M.K.; Chaturvedi, R.; Chowdhury, Z.; Shah, J.; Nandi, A.K. Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol. Plant Microbe Interact. 2013, 26, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006, 141, 1666–1675. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.; Hennig, J.; Klessig, D.F. Temperature-dependent induction of salicylic acid and its conjugates during the resistance response to tobacco mosaic virus infection. Plant Cell 1992, 4, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.I.; Raskin, I. Purification, cloning, and expression of a pathogen inducible UDP-glucose: Salicylic acid glucosyltransferase from tobacco. J. Biol. Chem. 1999, 274, 36637–36642. [Google Scholar] [CrossRef] [PubMed]
- Park, S.W.; Liu, P.P.; Forouhar, F.; Vlot, A.C.; Tong, L.; Tietjen, K.; Klessig, D.F. Use of a synthetic salicylic acid analog to investigate the roles of methyl salicylate and its esterases in plant disease resistance. J. Biol. Chem. 2009, 284, 7307–7317. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Klessig, D.F.; Park, S.W. Systemic acquired resistance: The elusive signal(s). Curr. Opin. Plant Biol. 2008, 11, 436–442. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Liu, P.P.; Cameron, R.K.; Park, S.W.; Yang, Y.; Kumar, D.; Zhou, F.; Padukkavidana, T.; Gustafsson, C.; Pichersky, E.; et al. Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 2008, 56, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, D.A.; Klessig, D.F. SOS—Too many signals for systemic acquired resistance? Trends Plant Sci. 2012, 17, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; von Dahl, C.C.; Klessig, D.F. The extent to which methyl salicylate is required for signaling systemic acquired resistance is dependent on exposure to light after infection. Plant Physiol. 2011, 157, 2216–2226. [Google Scholar] [CrossRef] [PubMed]
- Guedes, M.E.M.; Richmond, S.; Kuć, J. Induced systemic resistance to anthracnose in cucumber as influenced by the location of the inducer inoculation with Colletotrichum lagenarium and the onset of flowering and fruiting. Physiol. Plant Pathol. 1980, 17, 229–233. [Google Scholar] [CrossRef]
- Van Bel, A.J.E.; Gaupels, F. Pathogen-induced resistance and alarm signals in the phloem. Mol. Plant Pathol. 2004, 5, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Riedlmeier, M.; Ghirardo, A.; Wenig, M.; Knappe, C.; Koch, K.; Georgii, R.; Dey, S.; Parker, J.E.; Schnitzler, J.P.; Vlot, A.C. Monoterpenes support systemic acquired resistance within and between plants. Plant Cell 2017, 29, 1440–1459. [Google Scholar] [CrossRef] [PubMed]
- Miquel, M.; Cassagne, C.; Browse, J. A new class of Arabidopsis mutants with reduced hexadecatrienoic acid fatty acid levels. Plant Physiol. 1998, 117, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Lorenc-Kukula, K.; Chaturvedi, R.; Roth, M.; Welti, R.; Shah, J. Biochemical and molecular-genetic characterization of SFD1’s involvement in lipid metabolism and defense signaling. Front. Plant Sci. 2012, 3, 26. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, R.; Krothapalli, K.; Makandar, R.; Nandi, A.; Sparks, A.A.; Roth, M.R.; Welti, R.; Shah, J. Plastid omega3-fatty acid desaturase-dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 2008, 54, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.M.; Yu, K.; Xia, Y.; Shine, M.B.; Wang, C.; Navarre, D.; Kachroo, A.; Kachroo, P. Mono- and digalactosyldiacylglycerol lipids function nonredundantly to regulate systemic acquired resistance in plants. Cell Rep. 2014, 9, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Draelos, Z. Skin lightening preparations and the hydroquinone controversy. Dermatol. Ther. 2007, 20, 308–313. [Google Scholar] [CrossRef] [PubMed]
- Zoeller, M.; Stingl, N.; Krischke, M.; Fekete, A.; Waller, F.; Berger, S.; Mueller, M.J. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: Biogenesis of pimelic and azelaic acid. Plant Physiol. 2012, 160, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Wittek, F.; Hoffmann, T.; Kanawati, B.; Bichlmeier, M.; Knappe, C.; Wenig, M.; Schmitt-Kopplin, P.; Parker, J.E.; Schwab, W.; Vlot, A.C. Arabidopsis ENHANCED DISEASE SUSCEPTIBILITY1 promotes systemic acquired resistence via azelaic acid and its precursor 9-oxo nonanoic acid. J. Exp. Bot. 2014, 65, 5919–5931. [Google Scholar] [CrossRef] [PubMed]
- Djami-Tchatchou, A.T.; Ncube, E.N.; Steenkamp, P.A.; Dubery, I.A. Similar, but different: Structurally related azelaic acid and hexanoic acid trigger differential metabolomic and transcriptomic responses in tobacco cells. BMC Plant Biol. 2017, 17, 227. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- Vicente, J.; Cascon, T.; Vicedo, B.; Garcia-Agustin, P.; Hamberg, M.; Castresana, C. Role of 9-lipoxygenase and α-dioxygenase oxylipin pathways as modulators of local and systemic defense. Mol. Plant 2012, 5, 914–928. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; El-Shetehy, M.; Shine, M.B.; Yu, K.; Navarre, D.; Wendehenne, D.; Kachroo, A.; Kachroo, P. Free radicals mediate systemic acquired resistance. Cell Rep. 2014, 7, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.; Chaturvedi, R.; Chowdhury, Z.; Venables, B.; Petros, R.A. Signaling by small metabolites in systemic acquired resistance. Plant J. 2014, 79, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Hamberger, B.; Ohnishi, T.; Hamberger, B.; Séguin, A.; Bohlmann, J. Evolution of diterpene metabolism: Sitka spruce CYP720B4 catalyzes multiple oxidations in resin acid biosynthesis of conifer defense against insects. Plant Physiol. 2011, 157, 1677–1695. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Sung, S. Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Curr. Opin. Plant Biol. 2012, 15, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Roy, S.; Singh, D.; Nandi, A.K. Arabidopsis FLOWERING LOCUS D influences systemic-acquired-resistance- induced expression and histone modifications of WRKY genes. J. Biosci. 2014, 39, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Plecko, B.; Hikel, C.; Korenke, G.C. Pipecolic acid as a diagnostic marker of pyridoxine-dependent epilepsy. Neuropediatrics 2005, 36, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Tranchant, C.; Aubourg, P.; Mohr, M.; Rocchiccioli, F.; Zaenker, C.; Warter, J.M. A new peroxisomal disease with impaired phytanic and pipecolic acid oxidation. Neurology 1993, 43, 2044–2048. [Google Scholar] [CrossRef] [PubMed]
- Kvenholden, K.A.; Lawless, J.G.; Ponnamperuma, C. Nonprotein amino acids in the murchison meteorite. Proc. Natl. Acad. Sci. USA. 1971, 68, 486–490. [Google Scholar] [CrossRef]
- Pálfi, G.; Dézsi, L. Pipecolic acid as an indicator of abnormal protein metabolism in diseased plants. Plant Soil 1968, 29, 285–291. [Google Scholar] [CrossRef]
- Song, J.T.; Lu, H.; Greenberg, J.T. Divergent roles in Arabidopsis thaliana development and defense of two homologous genes, ABERRANT GROWTH AND DEATH2 and AGD2-LIKE DEFENSE RESPONSE PROTRIN1, encoding novel aminotransferases. Plant Cell 2004, 16, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Ding, P.; Rekhter, D.; Ding, Y.; Feussner, K.; Busta, L.; Haroth, S.; Xu, S.; Li, X.; Jetter, R.; Feussner, I.; et al. Characterization of a pipecolic acid biosynthesis pathway required for systemic acquired resistance. Plant Cell 2016, 28, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Busta, L.; Zhang, Q.; Ding, P.; Jetter, R.; Zhang, Y. TGAGG-BINDING FACTOR1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol. 2018, 217, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin monooxygenase-generated N-hydroxypipecolic acid is a critical element of plant systemic immunity. Cell 2018, 17, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Holmes, E.C.; Rajniak, J.; Kim, J.-G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S.; Stanford University, Stanford, CA, USA. N-hydroxy-pipecolic acid is a mobile signal that induces systemic disease resistance in Arabidopsis. 2018. Preprint. Available online: https://www.biorxiv.org/content/early/2018/03/25/288449 (accessed on 9 April 2018).
- Zeier, J.; Pink, B.; Mueller, M.J.; Berger, S. Light conditions influence specific defence responses in incompatible plant-pathogen interactions: Uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 2004, 219, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Griebel, T.; Zeier, J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008, 147, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Genoud, T.; Buchala, A.J.; Chua, N.H.; Metraux, J.P. Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis. Plant J. 2002, 31, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Muhlenbock, P.; Szechynska-Hebda, M.; Plaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpinska, B.; Karpinski, S. Chloroplast signaling and lesion simulating disease1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef] [PubMed]
- Fodor, J.; Gullner, G.; Ádám, A.L.; Barna, B.; Kőmíves, T.; Király, Z. Local and systemic responses of antioxidants to tobacco mosaic virus infection and to salicylic acid in tobacco (Role in systemic acquired resistance). Plant Physiol. 1997, 114, 1443–1451. [Google Scholar] [CrossRef] [PubMed]
- Chandra-Shekara, A.C.; Gupte, M.; Navarre, D.; Raina, S.; Raina, R.; Klessig, D.; Kachroo, P. Light-dependent hypersensitive response and resistance signaling against Turnip Crinkle Virus in Arabidopsis. Plant J. 2006, 45, 320–334. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ádám, A.L.; Nagy, Z.Á.; Kátay, G.; Mergenthaler, E.; Viczián, O. Signals of Systemic Immunity in Plants: Progress and Open Questions. Int. J. Mol. Sci. 2018, 19, 1146. https://doi.org/10.3390/ijms19041146
Ádám AL, Nagy ZÁ, Kátay G, Mergenthaler E, Viczián O. Signals of Systemic Immunity in Plants: Progress and Open Questions. International Journal of Molecular Sciences. 2018; 19(4):1146. https://doi.org/10.3390/ijms19041146
Chicago/Turabian StyleÁdám, Attila L., Zoltán Á. Nagy, György Kátay, Emese Mergenthaler, and Orsolya Viczián. 2018. "Signals of Systemic Immunity in Plants: Progress and Open Questions" International Journal of Molecular Sciences 19, no. 4: 1146. https://doi.org/10.3390/ijms19041146