Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits
Abstract
:1. Introduction
2. Results and Discussion
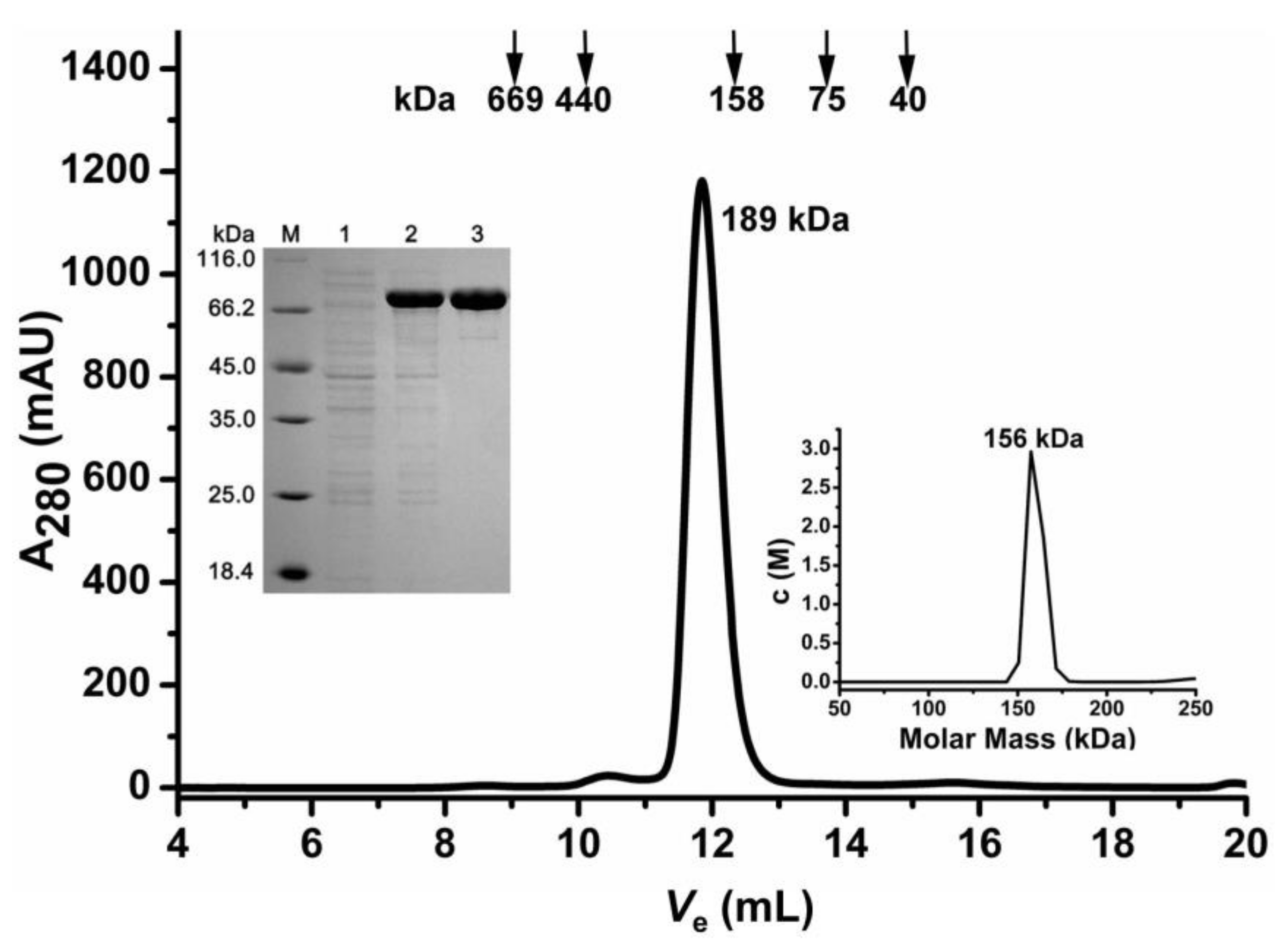
2.1. Oligomeric State Determination of the Recombinant AbIDH2
2.2. Kinetic Characterization of the Recombinant AbIDH2
2.3. Crystal Structure of the Dimeric AbIDH2
2.4. Mutational Analysis of the AbIDH2
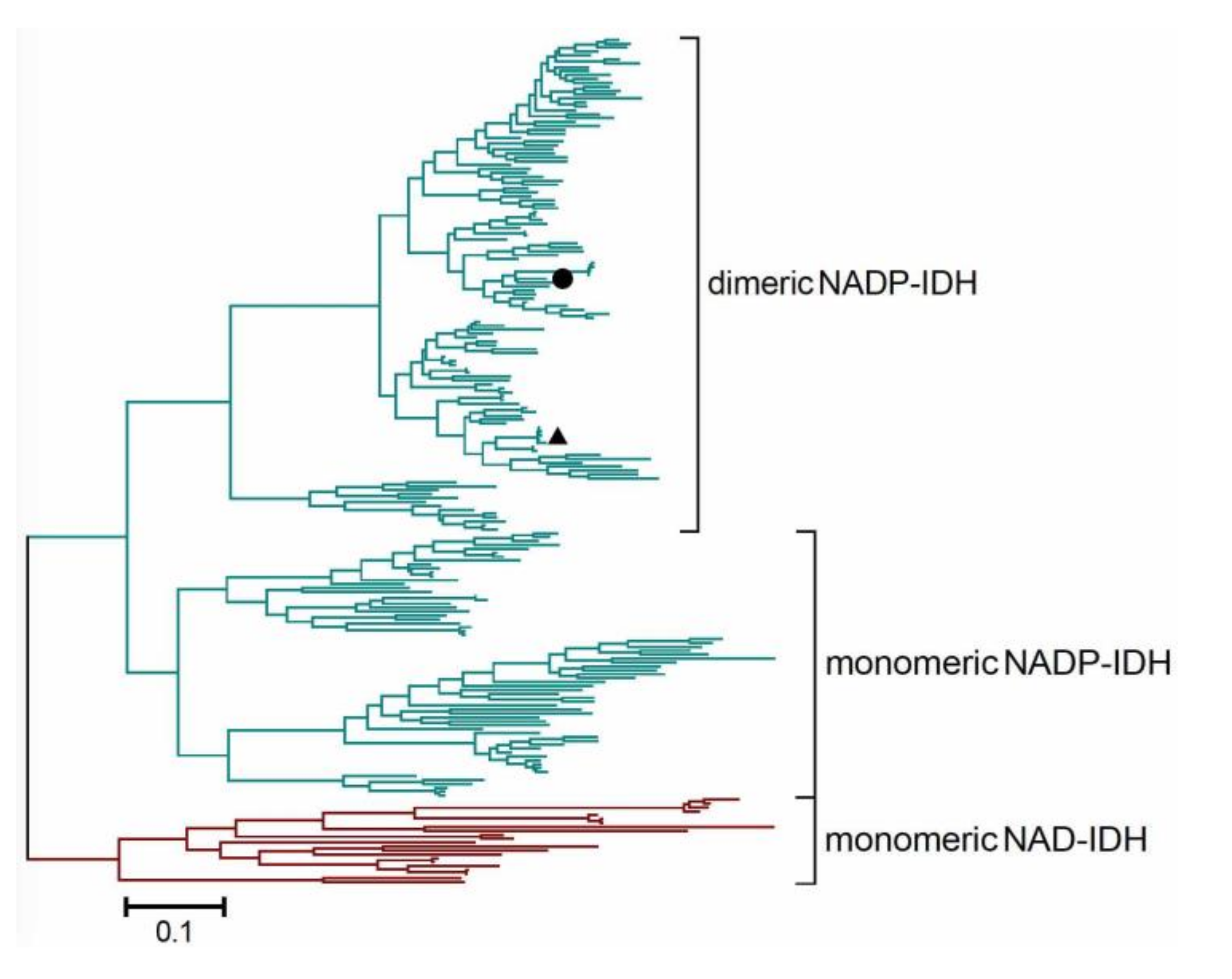
2.5. Redefining the Phylogenesis of the Monomeric IDH Subfamily
3. Materials and Methods
3.1. Cloning, Expression, and Purification of AbIDH2
3.2. SEC Chromatography
3.3. Sedimentation Velocity
3.4. Enzyme Assay and Kinetic Determination
3.5. Crystallization and Structure Determination
3.6. Sequence Alignments and Phylogenetic Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Taylor, A.B.; Hu, G.; Hart, P.J.; McAlister-Henn, L. Allosteric motions in structures of yeast NAD+-specific isocitrate dehydrogenase. J. Biol. Chem. 2008, 283, 10872–10880. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Golding, G.B.; Dean, A.M. The selective cause of an ancient adaptation. Science 2005, 307, 1279–1282. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.H.; Son, M.K.; Koh, H.J.; Lee, S.M.; Song, I.H.; Kim, Y.O.; Lee, Y.S.; Jeong, K.S.; Kim, W.B.; Park, J.W.; et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001, 276, 16168–16176. [Google Scholar] [CrossRef] [PubMed]
- Imabayashi, F.; Aich, S.; Prasad, L.; Delbaere, L.T. Substrate-free structure of a monomeric NADP isocitrate dehydrogenase: An open conformation phylogenetic relationship of isocitrate dehydrogenase. Proteins 2006, 63, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Stokke, R.; Madern, D.; Fedoy, A.E.; Karlsen, S.; Birkeland, N.K.; Steen, I.H. Biochemical characterization of isocitrate dehydrogenase from Methylococcus capsulatus reveals a unique NAD+-dependent homotetrameric enzyme. Arch. Microbiol. 2007, 187, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.M.; Golding, G.B. Protein engineering reveals ancient adaptive replacements in isocitrate dehydrogenase. Proc. Natl. Acad. Sci. USA 1997, 94, 3104–3109. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Lv, C.; Zhu, G. Novel type II and monomeric NAD+ specific isocitrate dehydrogenases: Phylogenetic affinity, enzymatic characterization, and evolutionary implication. Sci. Rep. 2015, 5, 9150. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.C.; Tian, C.Q.; Cheng, H.M.; Xu, L.; Wang, P.; Zhu, G.P. A novel type II NAD+-specific isocitrate dehydrogenase from the marine bacterium Congregibacter litoralis KT71. PLoS ONE 2015, 10, e0125229. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.G.; Song, P.; Cao, Z.Y.; Wang, P.; Zhu, G.P. A unique homodimeric NAD+-linked isocitrate dehydrogenase from the smallest autotrophic eukaryote Ostreococcus tauri. FASEB J. 2015, 29, 2462–2472. [Google Scholar] [CrossRef] [PubMed]
- Yasutake, Y.; Watanabe, S.; Yao, M.; Takada, Y.; Fukunaga, N.; Tanaka, I. Crystal structure of the monomeric isocitrate dehydrogenase in the presence of NADP+: Insight into the cofactor recognition, catalysis, and evolution. J. Biol. Chem. 2003, 278, 36897–36904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, B.; Wang, P.; Cao, Z.; Huang, E.; Hao, J.; Dean, A.M.; Zhu, G. Enzymatic characterization of a monomeric isocitrate dehydrogenase from Streptomyces lividans TK54. Biochimie 2009, 91, 1405–1410. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Nandyala, A.; Podili, R.; Katoch, V.M.; Hasnain, S.E. Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem. 2005, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Folta-Stogniew, E.; Williams, K.R. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Technol. 1999, 10, 51–63. [Google Scholar]
- Arakawa, T.; Ejima, D.; Li, T.; Philo, J.S. The critical role of mobile phase composition in size exclusion chromatography of protein pharmaceuticals. J. Pharm. Sci. 2010, 99, 1674–1692. [Google Scholar] [CrossRef] [PubMed]
- Sahara, T.; Takada, Y.; Takeuchi, Y.; Yamaoka, N.; Fukunaga, N. Cloning, sequencing, and expression of a gene encoding the monomeric isocitrate dehydrogenase of the nitrogen-fixing bacterium, Azotobacter vinelandii. Biosci. Biotechnol. Biochem. 2002, 66, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yang, H. A highly specific monomeric isocitrate dehydrogenase from Corynebacterium glutamicum. Arch. Biochem. Biophys. 2000, 383, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Greer, A.; Dean, A.M. A highly active decarboxylating dehydrogenase with rationally inverted coenzyme specificity. Proc. Natl. Acad. Sci. USA 1995, 92, 11666–11670. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Li, S.; Wu, Y.; Lv, C.; Wang, P.; Zhu, G. Point mutation (R153H or R153C) in Escherichia coli isocitrate dehydrogenase: Biochemical characterization and functional implication. J. Basic Microbiol. 2017, 57, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Yasutake, Y.; Watanabe, S.; Yao, M.; Takada, Y.; Fukunaga, N.; Tanaka, I. Structure of the monomeric isocitrate dehydrogenase: Evidence of a protein monomerization by a domain duplication. Structure 2002, 10, 1637–1648. [Google Scholar] [CrossRef]
- Watanabe, S.; Yasutake, Y.; Tanaka, I.; Takada, Y. Elucidation of stability determinants of cold-adapted monomeric isocitrate dehydrogenase from a psychrophilic bacterium, Colwellia maris, by construction of chimeric enzymes. Microbiology 2005, 151, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, N.S.; Delbaere, L.T.; Sheldrick, G.M. Structure of a highly NADP+-specific isocitrate dehydrogenase. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Cao, Z.Y.; Wang, P.; Liu, A.M.; Pan, W.; Wang, J.; Zhu, G.P. Heteroexpression and characterization of a monomeric isocitrate dehydrogenase from the multicellular prokaryote Streptomyces avermitilis MA-4680. Mol. Biol. Rep. 2011, 38, 3717–3724. [Google Scholar] [CrossRef] [PubMed]
- Lv, C.; Wang, P.; Wang, W.; Su, R.; Ge, Y.; Zhu, Y.; Zhu, G. Two isocitrate dehydrogenases from a plant pathogen Xanthomonas campestris pv. campestris 8004. Bioinformatic analysis, enzymatic characterization, and implication in virulence. J. Basic Microbiol. 2016, 56, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Doughari, H.J.; Ndakidemi, P.A.; Human, I.S.; Benade, S. The ecology, biology and pathogenesis of Acinetobacter spp.: An overview. Microb. Environ. 2011, 26, 101–112. [Google Scholar] [CrossRef]
- Roca, I.; Espinal, P.; Vila-Farres, X.; Vila, J. The Acinetobacter baumannii oxymoron: Commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 2012, 3, 148. [Google Scholar] [CrossRef] [PubMed]
- Stafford, W.F., III. Boundary analysis in sedimentation transport experiments: A procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992, 203, 295–301. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]


| Enzyme | Isocitrate | NADP+ | ||||
|---|---|---|---|---|---|---|
| Km (μM) | kcat (s−1) | kcat/Km (μM−1·s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | |
| AbIDH2 (Mg2+) | 21 ± 4 | 5.2 ± 0.3 | 0.24 | 159 ± 23 | 7.4 ± 0.6 | 0.05 |
| AbIDH2 (Mn2+)5 | 21 ± 3 | 39.2 ± 2.1 | 1.9 | 94 ± 6 | 36.9 ± 1.2 | 0.39 |
| Statistics | AbIDH2 | |
|---|---|---|
| Data Collection | Space group | P4 222 |
| Cell dimensions | ||
| a, b, c (Å) | 137.16, 137.16, 238.13 | |
| α, β, γ (°) | 90.00, 90.00, 90.00 | |
| Wavelength (Å) | 0.97776 | |
| Resolution (Å) a | 48.54–3.0 (3.05–3.0) | |
| Rsym or Rmerge | 0.132 (0.402) | |
| Average I/σ(I) | 3.2 (1.96) | |
| Completeness (%) | 99.96 (100) | |
| Redundancy | 3.3 (3.2) | |
| Refinement | Resolution (Å) | 3.0 |
| No. reflections | 46,278 (4541) | |
| Rwork/Rfree | 0.205 (0.245)/0.276 (0.323) | |
| No. atoms | ||
| Protein | 11,408 | |
| Ligand/ion | 12 | |
| Water | 369 | |
| B-factors | ||
| Protein | 54.2 | |
| Ligand/ion | 48.7 | |
| Water | 57.2 | |
| R.m.s deviations b | ||
| Bond lengths (Å) | 0.0114 | |
| Bond angles (°) | 1.688 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Wu, Y.; Liu, J.; Song, P.; Li, S.; Zhou, X.; Zhu, G. Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits. Int. J. Mol. Sci. 2018, 19, 1131. https://doi.org/10.3390/ijms19041131
Wang P, Wu Y, Liu J, Song P, Li S, Zhou X, Zhu G. Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits. International Journal of Molecular Sciences. 2018; 19(4):1131. https://doi.org/10.3390/ijms19041131
Chicago/Turabian StyleWang, Peng, Yatao Wu, Jie Liu, Ping Song, Shan Li, Xinxin Zhou, and Guoping Zhu. 2018. "Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits" International Journal of Molecular Sciences 19, no. 4: 1131. https://doi.org/10.3390/ijms19041131




