miR-146b Inhibits Glucose Consumption by Targeting IRS1 Gene in Porcine Primary Adipocytes
Abstract
:1. Introduction
2. Results
2.1. miR-146b Inhibits Glucose Consumptionin
2.2. Target Prediction and Pathway Analysis
2.3. miR-146b Repressed GLUT4 and IRS1 Protein Expression
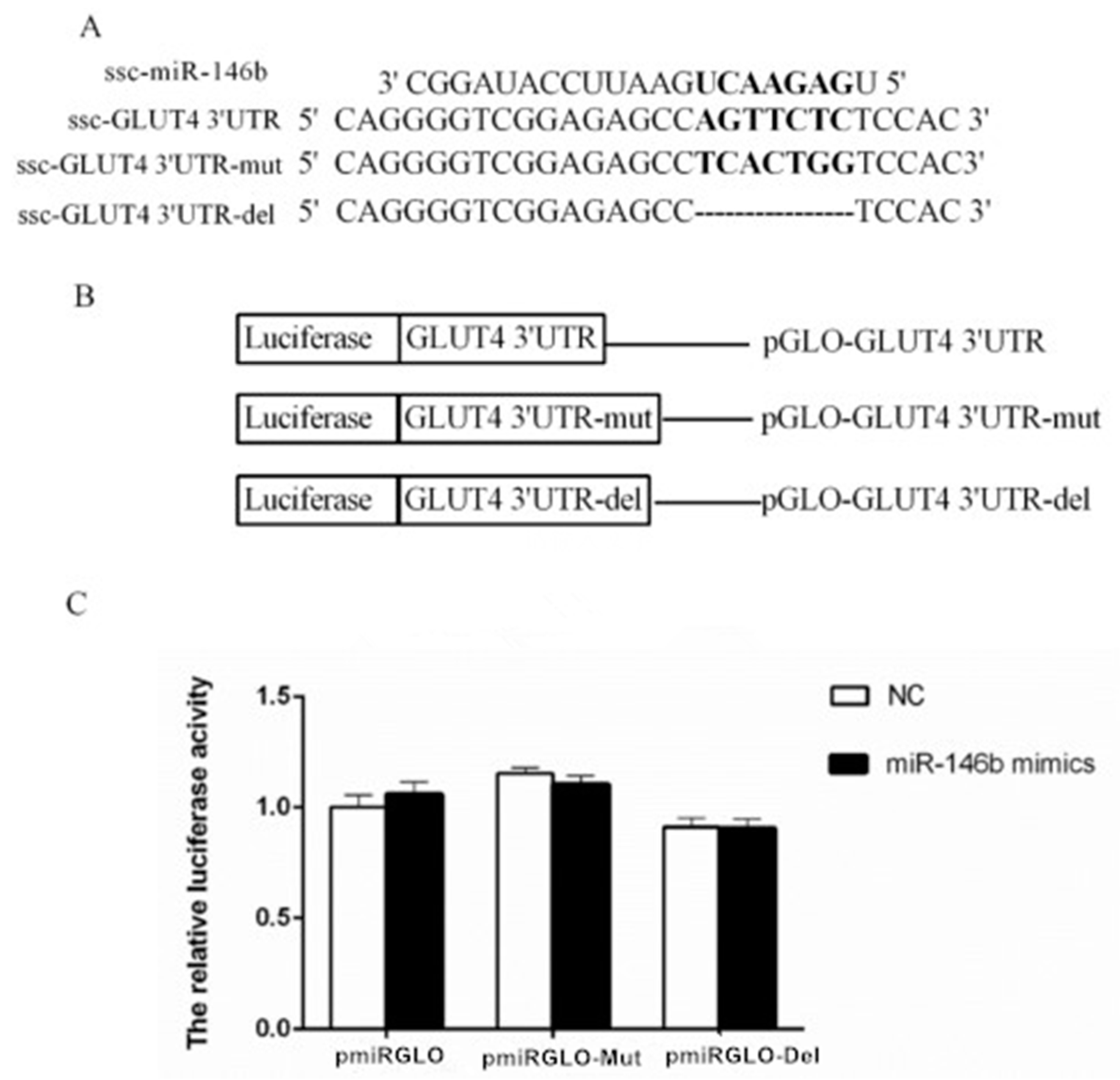
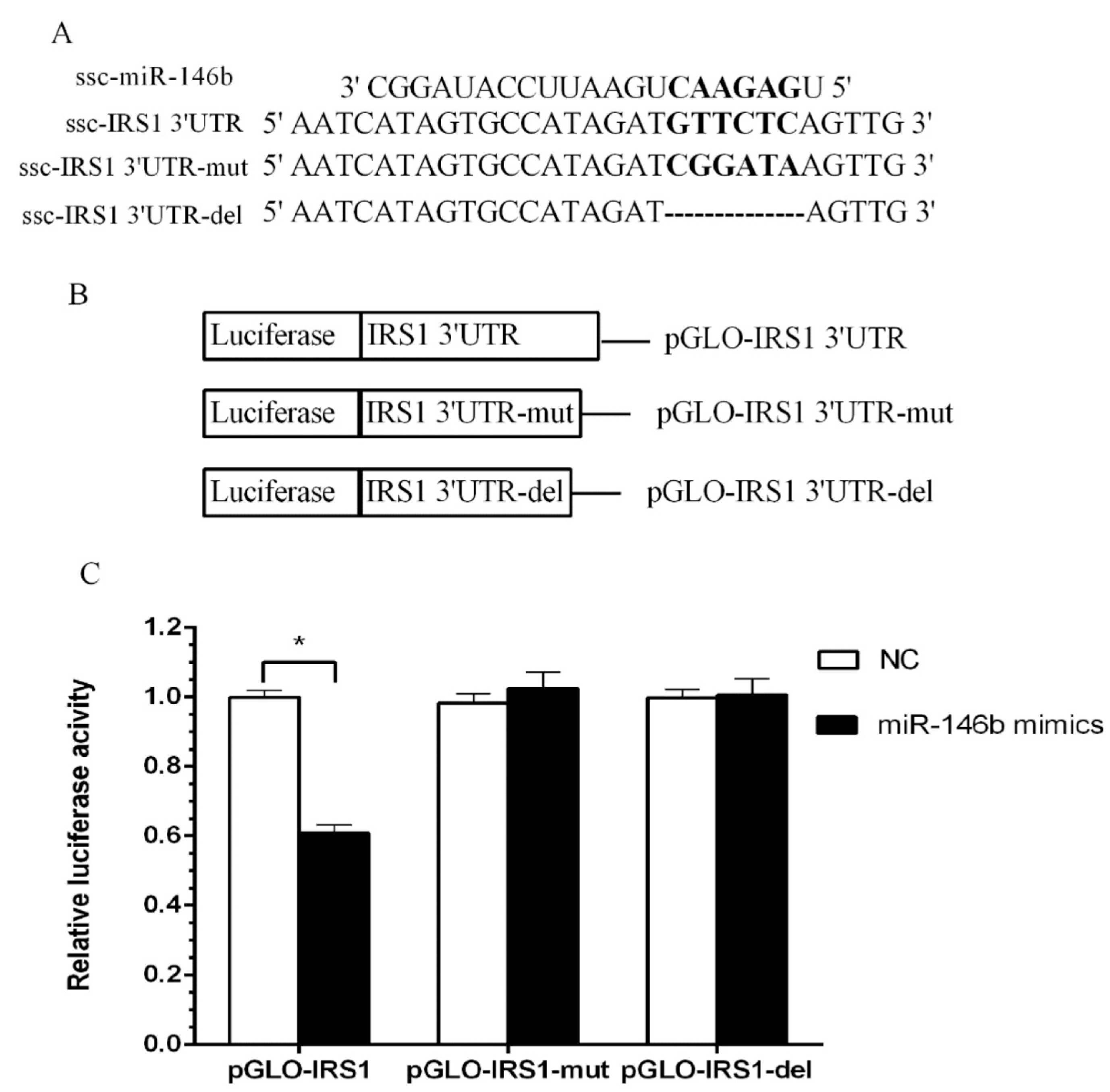
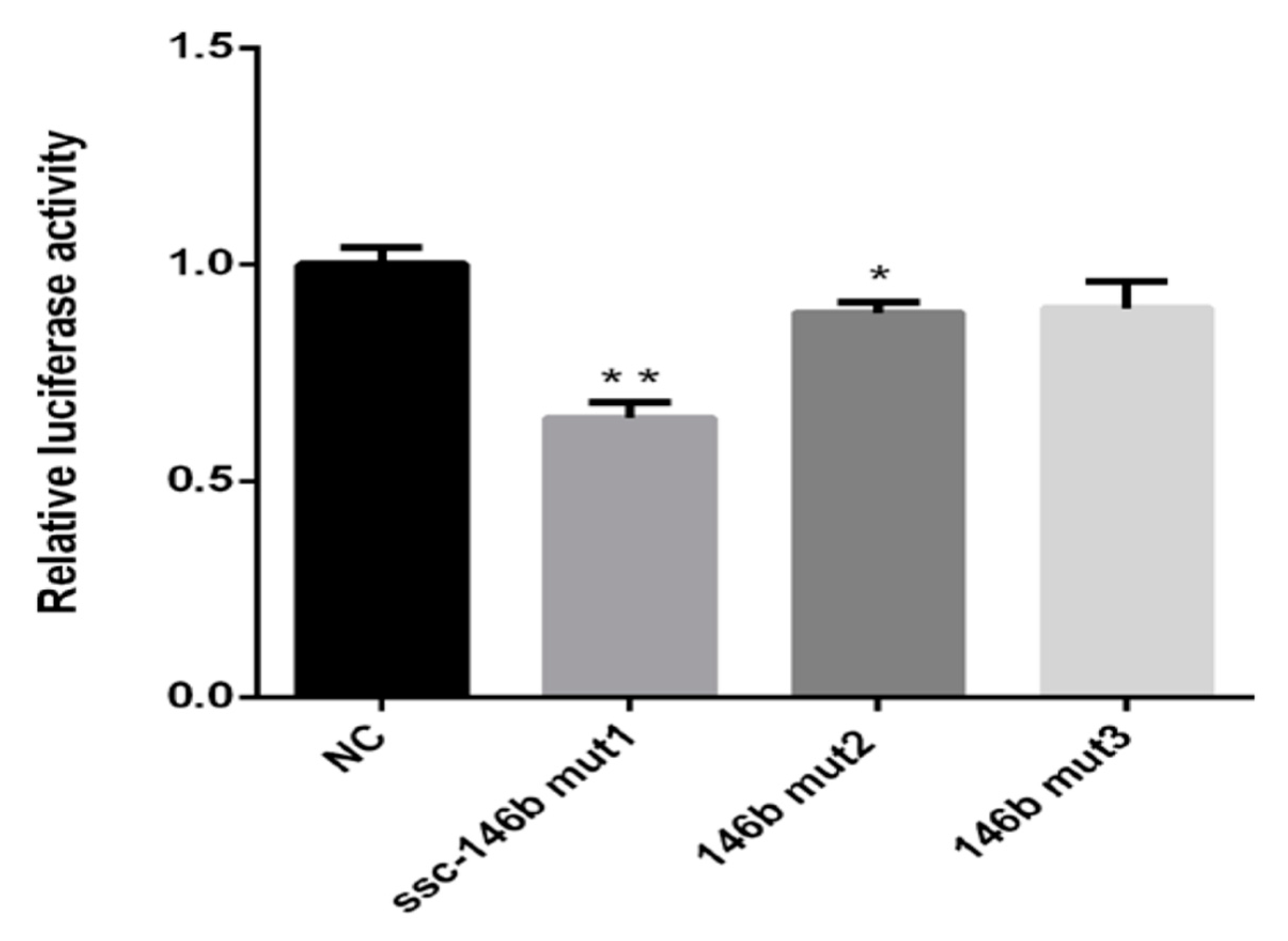
2.4. Identification of miR-146b Targets by Luciferase Reporter Assay
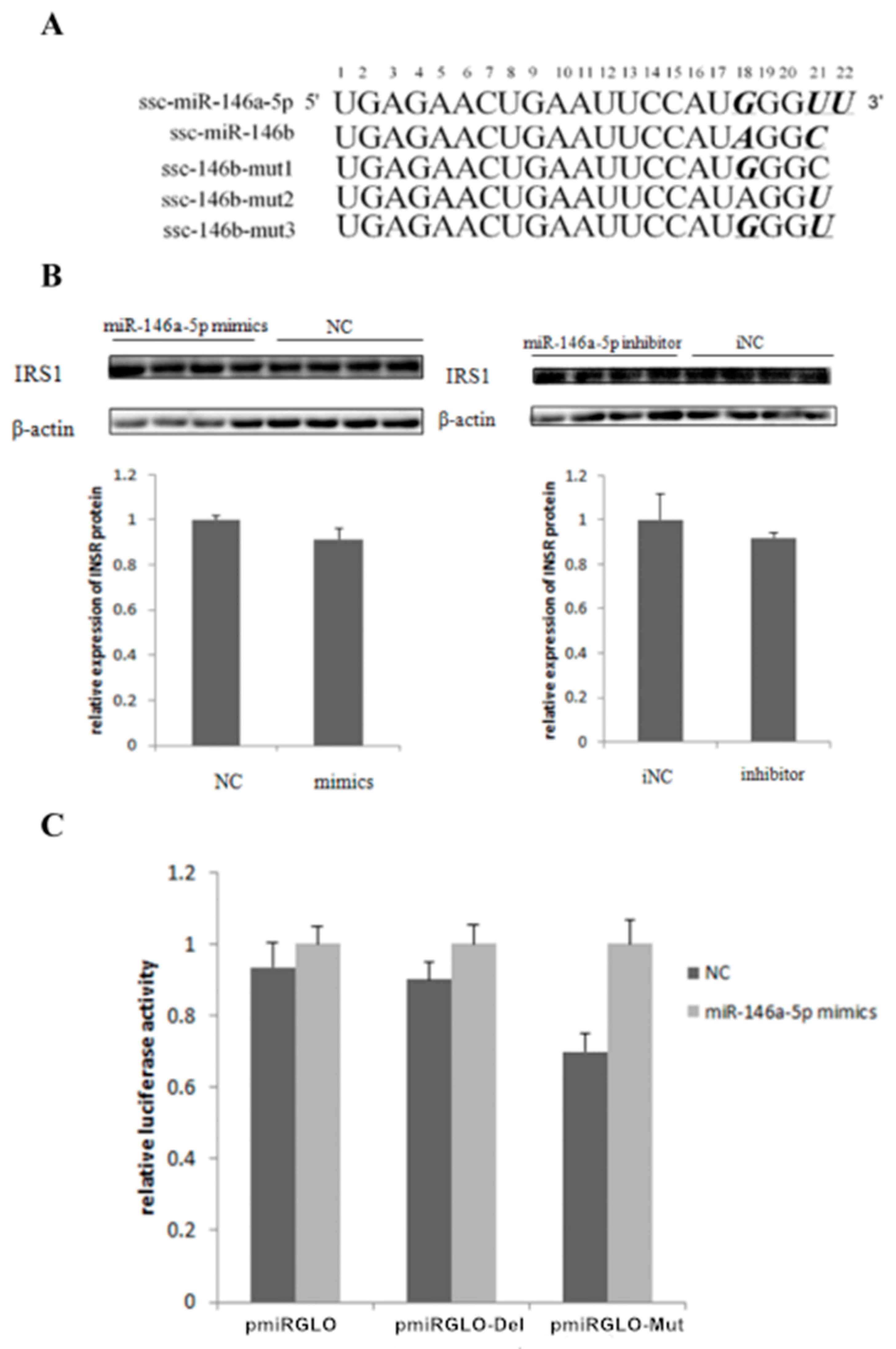
2.5. miR-146bend Base Mutation Changes miR-146b Targeting on IRS1 by Luciferase Reporter Assay
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection and Culture of Porcinepre-Adipocytes
4.3. Target Prediction and Pathway Analysis
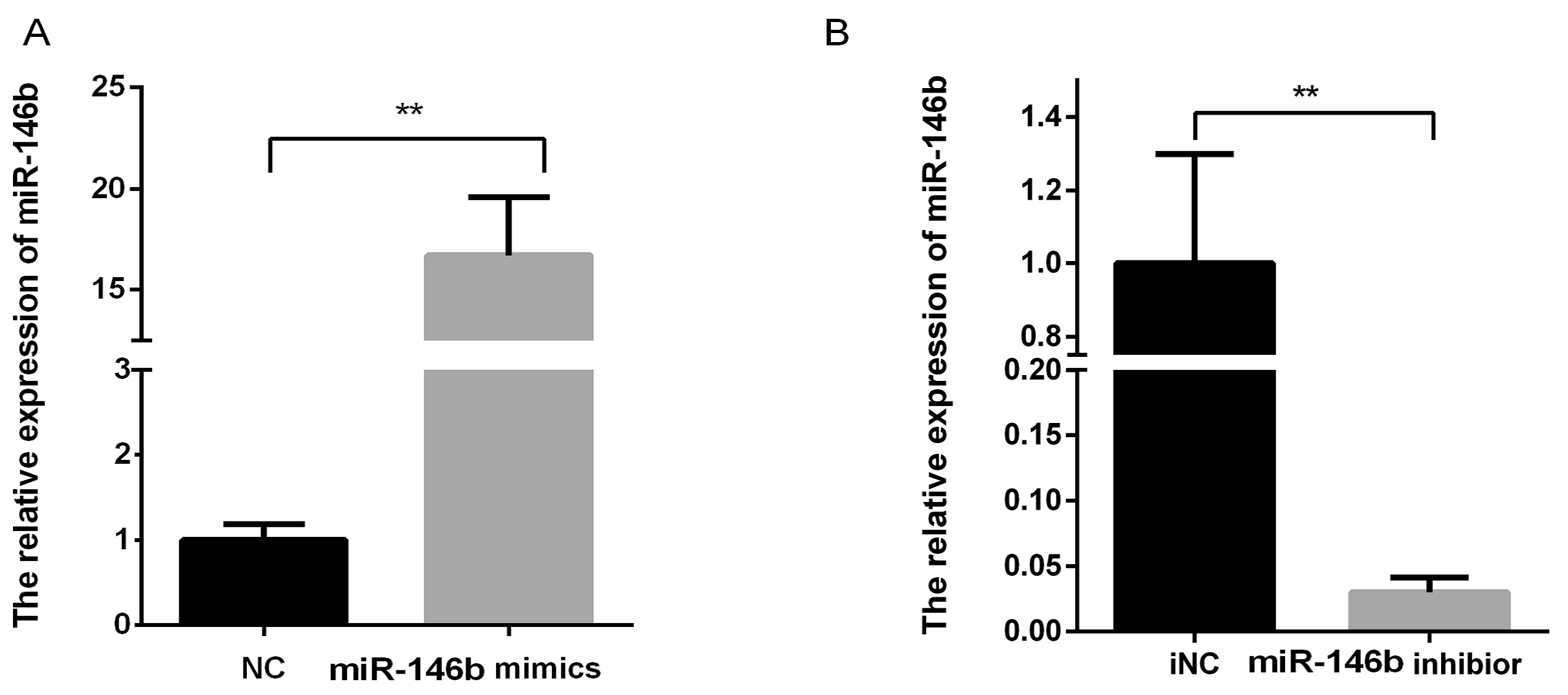
4.4. Transfection of miR-146b Mimics and miR-146b Inhibitor
4.5. RNA Extraction and Real-Time PCR for miR-146b
4.6. Glucose Consumption Assay
4.7. Protein Extraction and Western Blot
4.8. Plasmid Construction
4.9. Dual-Luciferase Reporter Assay
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fantuzzi, G. Adipose tissue, adipokines, and inflammation. J. Allergy Clin. Immunol. 2005, 115, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Bozaoglu, K.; Bolton, K.; McMillan, J.; Zimmet, P.; Jowett, J.; Collier, G.; Walder, K.; Segal, D. Chemerin is a novel adipokine associated with obesity and metabolic syndrome. Endocrinology 2007, 148, 4687–4694. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Chehab, F.F.; Mounzih, K.; Lu, R.; Lim, M.E. Early onset of reproductive function in normal female mice treated with leptin. Science 1997, 275, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Mokdad, A.H.; Ford, E.S.; Bowman, B.A.; Dietz, W.H.; Vinicor, F.; Bales, V.S.; Marks, J.S. Prevalence of obesity, diabetes, and obesity-related health risk factors, 2001. JAMA 2003, 289, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Herman, M.A.; Kahn, B.B. Glucose transport and sensing in the maintenance of glucose homeostasis and metabolic harmony. J. Clin. Investig. 2006, 116, 1767–1775. [Google Scholar] [CrossRef] [PubMed]
- Tirone, T.A.; Brunicardi, F.C. Overview of Glucose Regulation. World J. Surg. 2001, 25, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. Adipocytes as regulators of energy balance and glucose homeostasis. Nature 2006, 444, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Lunney, J.K. Advances in Swine Biomedical Model Genomics. Pigs Poult. 2008, 3, 179–184. [Google Scholar] [CrossRef]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Rep. 2001, 2, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Kono, T. Evidence That Insulin Causes Translocation of Glucose Transport Activity to the Plasma Membrane from an Intracellular Storage Site. Proc. Natl. Acad. Sci. USA 1980, 77, 2542–2545. [Google Scholar] [CrossRef] [PubMed]
- Wardzala, L.J.; Jeanrenaud, B. Potential mechanism of insulin action on glucose transport in the isolated rat diaphragm. Apparent translocation of intracellular transport units to the plasma membrane. J. Biol. Chem. 1981, 256, 7090–7093. [Google Scholar] [PubMed]
- Shepherd, P.R.; Kahn, B.B. Glucose transporters and insulin action—Implications for insulin resistance and diabetes mellitus. N. Engl. J. Med. 1999, 341, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Stenbit, A.E.; Tsao, T.S.; Li, J.; Burcelin, R.; Geenen, D.L.; Factor, S.M.; Houseknecht, K.; Katz, E.B.; Charron, M.J. GLUT4 heterozygous knockout mice develop muscle insulin resistance and diabetes. Nat. Med. 1997, 3, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Brozinick, J.T., Jr.; McCoid, S.C.; Reynolds, T.H.; Nardone, N.A.; Hargrove, D.M.; Stevenson, R.W.; Cushman, S.W.; Gibbs, E.M. GLUT4 overexpression in db/db mice dose-dependently ameliorates diabetes but is not a lifelong cure. Diabetes 2001, 50, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Biochemistry, 5th ed.; W H Freeman: New York, NY, USA, 2002. [Google Scholar]
- Martin, S.; Tellam, J.; Livingstone, C.; Slot, J.W.; Gould, G.W.; James, D.E. The glucose transporter (GLUT-4) and vesicle-associated membrane protein-2 (VAMP-2) are segregated from recycling endosomes in insulin-sensitive cells. J. Cell Biol. 1996, 134, 625–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slot, J.W.; Geuze, H.J.; Gigengack, S.; Lienhard, G.E.; James, D.E. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 1991, 113, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Millar, C.A.; Lyttle, C.T.; Meerloo, T.; Marsh, B.J.; Gould, G.W.; James, D.E. Effects of insulin on intracellular GLUT4 vesicles in adipocytes: Evidence for a secretory mode of regulation. J. Cell Sci. 2000, 113 Pt 19, 3427–3438. [Google Scholar] [PubMed]
- Cheng, Z.; Tseng, Y.; White, M.F. Insulin signaling meets mitochondria in metabolism. Trends Endocrinol. Metab. 2010, 21, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.F.; Ramm, G.; Larance, M.; Hoehn, K.L.; Wagner, M.C.; Guilhaus, M.; James, D.E. CaMKII-Mediated Phosphorylation of the Myosin Motor Myo1c Is Required for Insulin-Stimulated GLUT4 Translocation in Adipocytes. Cell Metab. 2008, 8, 384–398. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G., Jr.; White, M.F. Insulin signal transduction and the IRS proteins. Annu. Rev. Pharmacol. 1996, 36, 615–658. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Pilch, P.F. The insulin receptor: Structure, function, and signaling. Am. J. Physiol. 1994, 266, C319–C334. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, G.; Sajan, M.P.; Kanoh, Y.; Standaert, M.L.; Quon, M.J.; Reed, B.C.; Dikic, I.; Farese, R.V. Glucose activates protein kinase C-zeta /lambda through proline-rich tyrosine kinase-2, extracellular signal-regulated kinase, and phospholipase D: A novel mechanism for activating glucose transporter translocation. J. Biol. Chem. 2001, 276, 35537–35545. [Google Scholar] [CrossRef] [PubMed]
- Whiteman, E.L.; Cho, H.; Birnbaum, M.J. Role of Akt/protein kinase B in metabolism. Trends Endocrinol. Metab. 2002, 13, 444–451. [Google Scholar] [CrossRef]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef] [PubMed]
- Foran, P.G.; Fletcher, L.M.; Oatey, P.B.; Mohammed, N.; Dolly, J.O.; Tavaré, J.M. Protein kinase B stimulates the translocation of GLUT4 but not GLUT1 or transferrin receptors in 3T3-L1 adipocytes by a pathway involving SNAP-23, synaptobrevin-2, and/or cellubrevin. J. Biol. Chem. 1999, 274, 28087–28095. [Google Scholar] [CrossRef] [PubMed]
- Hajduch, E.; Alessi, D.R.; Hemmings, B.A.; Hundal, H.S. Constitutive activation of protein kinase B alpha by membrane targeting promotes glucose and system A amino acid transport, protein synthesis, and inactivation of glycogen synthase kinase 3 in L6 muscle cells. Diabetes 1998, 47, 1006–1013. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.M.; Clark, S.F.; Tucker, D.F.; Birnbaum, M.J.; James, D.E.; Macaulay, S.L. A Role for Protein Kinase Bβ/Akt2 in Insulin-Stimulated GLUT4 Translocation in Adipocytes. Mol. Cell. Biol. 1999, 19, 7771–7781. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Croce, C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer 2006, 6, 857–866. [Google Scholar] [CrossRef] [PubMed]
- Romao, J.M.; Jin, W.; Dodson, M.V.; Hausman, G.J.; Moore, S.S.; Guan, L.L. MicroRNA regulation in mammalian adipogenesis. Exp. Biol. Med. 2011, 236, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, K.S.; Bae, H.J.; Eun, J.W.; Shen, Q.; Park, S.J.; Shin, W.C.; Yang, H.D.; Park, M.; Park, W.S.; et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget 2015, 6, 8089–8102. [Google Scholar] [CrossRef] [PubMed]
- Kohjima, M.; Higuchi, N.; Kato, M.; Kotoh, K.; Yoshimoto, T.; Fujino, T.; Yada, M.; Yada, R.; Harada, N.; Enjoji, M.; et al. SREBP-1c, regulated by the insulin and AMPK signaling pathways, plays a role in nonalcoholic fatty liver disease. Int. J. Mol. Med. 2008, 21, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Siddle, K. Signalling by insulin and IGF receptors: Supporting acts and new players. J. Mol. Endocrinol. 2011, 47, R1–R10. [Google Scholar] [CrossRef] [PubMed]
- Alexander, R.; Lodish, H.; Sun, L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin. Ther. Targets 2011, 15, 623–636. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; Dean, N.M.; et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [PubMed]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Sheedy, F.J.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van Gils, J.M.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in Mice Promotes Reverse Cholesterol Transport and Regression of Atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Zhu, L.; Gupta, N.; Chang, Y.; Fang, F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol. Endocrinol. 2007, 21, 2785–2794. [Google Scholar] [CrossRef] [PubMed]
- Leavens, K.F.; Birnbaum, M.J. Insulin signaling to hepatic lipid metabolism in health and disease. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Nandi, A.; Kitamura, Y.; Kahn, C.R.; Accili, D. Mouse models of insulin resistance. Physiol. Rev. 2004, 84, 623–647. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Karolina, D.S.; Armugam, A.; Tavintharan, S.; Wong, M.T.; Lim, S.C.; Sum, C.F.; Jeyaseelan, K. Correction: MicroRNA 144 Impairs Insulin Signaling by Inhibiting the Expression of Insulin Receptor Substrate 1 in Type 2 Diabetes Mellitus. PLoS ONE 2011, 6, e22839. [Google Scholar] [CrossRef]
- Yu, Y.; Li, X.; Liu, L.; Chai, J.; Haijun, Z.; Chu, W.; Yin, H.; Ma, L.; Duan, H.; Xiao, M. miR-628 Promotes Burn-Induced Skeletal Muscle Atrophy via Targeting IRS1. Int. J. Biol. Sci. 2016, 12, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Xi, Q.Y.; Cheng, X.; Dong, T.; Zhu, X.T.; Shu, G.; Wang, L.N.; Jiang, Q.Y.; Zhang, Y.L. miR-146a-5p inhibits TNF-α-induced adipogenesis via targeting insulin receptor in primary porcine adipocytes. J. Lipid Res. 2016, 57, 1360–1372. [Google Scholar] [CrossRef] [PubMed]
- Bryant, N.J.; Govers, R.; James, D.E. Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol. 2002, 3, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, M. Insulin receptor signals regulating GLUT4 translocation and actin dynamics. Endocr. J. 2006, 53, 267–293. [Google Scholar] [CrossRef] [PubMed]
- Haslam, D.W.; James, W.P. Obesity. Lancet 2005, 366, 1197–1209. [Google Scholar] [CrossRef]
- Cornelius, P.; MacDougald, O.A.; Lane, M.D. Regulation of adipocyte development. Annu. Rev. Nutr. 1994, 14, 99–129. [Google Scholar] [CrossRef] [PubMed]
- James, D.E.; Brown, R.; Navarro, J.; Pilch, P.F. Insulin-regulatable tissues express a unique insulin-sensitive glucose transport protein. Nature 1988, 333, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Brady, M.J.; Nairn, A.C.; Saltiel, A.R. The regulation of glycogen synthase by protein phosphatase 1 in 3T3-L1 adipocytes. Evidence for a potential role for DARPP-32 in insulin action. J. Biol. Chem. 1997, 272, 29698–29703. [Google Scholar] [CrossRef] [PubMed]
- White, M.F. IRS proteins and the common path to diabetes. AJP Endocrinol. Metab. 2002, 283, E413–E422. [Google Scholar] [CrossRef] [PubMed]
- Sesti, G.; Federici, M.; Hribal, M.L.; Lauro, D.; Sbraccia, P.; Lauro, R. Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J. 2001, 15, 2099–2111. [Google Scholar] [CrossRef] [PubMed]
- Smith, U. Impaired (‘diabetic’) insulin signaling and action occur in fat cells long before glucose intolerance—Is insulin resistance initiated in the adipose tissue? Int. J. Obes. Relat. Metab. Disord. 2002, 26, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Tanti, J.F.; Grémeaux, T.; van Obberghen, E.; Le Marchand-Brustel, Y. Serine/threonine phosphorylation of insulin receptor substrate 1 modulates insulin receptor signaling. J. Biol. Chem. 1994, 269, 6051–6057. [Google Scholar] [PubMed]
- Chassin, C.; Kocur, M.; Pott, J.; Duerr, C.U.; Gütle, D.; Lotz, M.; Hornef, M.W. miR-146a mediates protective innate immune tolerance in the neonate intestine. Cell Host Microbe 2010, 8, 358–368. [Google Scholar] [CrossRef] [PubMed]
- Curtale, G.; Mirolo, M.; Renzi, T.A.; Rossato, M.; Bazzoni, F.; Locati, M. Negative regulation of Toll-like receptor 4 signaling by IL-10–dependent microRNA-146b. Proc. Natl. Acad. Sci. USA 2013, 110, 11499–11504. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Sheedy, F.J.; O’Neill, L.A. Adding fuel to fire: MicroRNAs as a new class of mediators of inflammation. Ann. Rheum. Dis. 2008, 67, iii50–iii55. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Birkbak, N.J.; Vafaizadeh, V.; Walker, S.R.; Yeh, J.E.; Liu, S.; Kroll, Y.; Boldin, M.; Taganov, K.; Groner, B.; et al. STAT3 induction of miR-146b forms a feedback loop to inhibit the NF-κB to IL-6 signaling axis and STAT3-driven cancer phenotypes. Sci. Signal. 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Paterson, M.R.; Kriegel, A.J. miR-146a/b: A Family with Shared Seeds and Different Roots. Physiol. Genom. 2017, 49, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Trinder, P. Determination of blood glucose using an oxidase-peroxidase system with a non-carcinogenic chromogen. J. Clin. Pathol. 1969, 22, 158–161. [Google Scholar] [CrossRef] [PubMed]



| Pathway Term | Genes | p-Value |
|---|---|---|
| Adipocytokine signaling pathway | CPT1, FACS, IRS, AMPK, GLUT4, LEPR | 2.7 × 10−3 |
| T cell receptor signaling pathway | CD4/8, CD3y, Rho, Cdb42, NFAT, Ras, ICOS | 3.2 × 10−3 |
| Phagosome | MHCII, TLR4, Dynein, SRB1, TUBB, vATPase | 3.8 × 10−3 |
| Focal Adhesion | ECM, ITGA, ITGB, RhoA, MLC, Caveolin, Vinculin | 4.8 × 10−2 |
| Regulation of actin cytoskeleton | ITG, Ras, Rho, VCL, PI4P5K, MLC, Arp2/3, WAVE2 | 1.8 × 10−2 |
| Bacterial invasion of epithelial cells | WAVE, Arp2/3, RhoA, Vinculin, Caveollin | 2.0 × 10−2 |
| AMPK signaling pathway | Ob-Rb, AMPK, GLUT4, IRS1, Rab, CPT1 | 2.2 × 10−2 |
| FoxO signaling pathway | Smad4, AMPK, GLUT4, IRS, Ras, ATM | 3.5 × 10−2 |
| Neurotrophin Signaling Pathway | NT4, IRS1, Ras, RhoA, p53 | 7.7 × 10−2 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.-L.; Chen, T.; Xiong, J.-L.; Wu, D.; Xi, Q.-Y.; Luo, J.-Y.; Sun, J.-J.; Zhang, Y.-L. miR-146b Inhibits Glucose Consumption by Targeting IRS1 Gene in Porcine Primary Adipocytes. Int. J. Mol. Sci. 2018, 19, 783. https://doi.org/10.3390/ijms19030783
Zhu Y-L, Chen T, Xiong J-L, Wu D, Xi Q-Y, Luo J-Y, Sun J-J, Zhang Y-L. miR-146b Inhibits Glucose Consumption by Targeting IRS1 Gene in Porcine Primary Adipocytes. International Journal of Molecular Sciences. 2018; 19(3):783. https://doi.org/10.3390/ijms19030783
Chicago/Turabian StyleZhu, Yan-Ling, Ting Chen, Jia-Li Xiong, Di Wu, Qian-Yun Xi, Jun-Yi Luo, Jia-Jie Sun, and Yong-Liang Zhang. 2018. "miR-146b Inhibits Glucose Consumption by Targeting IRS1 Gene in Porcine Primary Adipocytes" International Journal of Molecular Sciences 19, no. 3: 783. https://doi.org/10.3390/ijms19030783





