iTRAQ-Based Proteomic Analysis Reveals Potential Regulation Networks of IBA-Induced Adventitious Root Formation in Apple
Abstract
:1. Introduction
2. Results
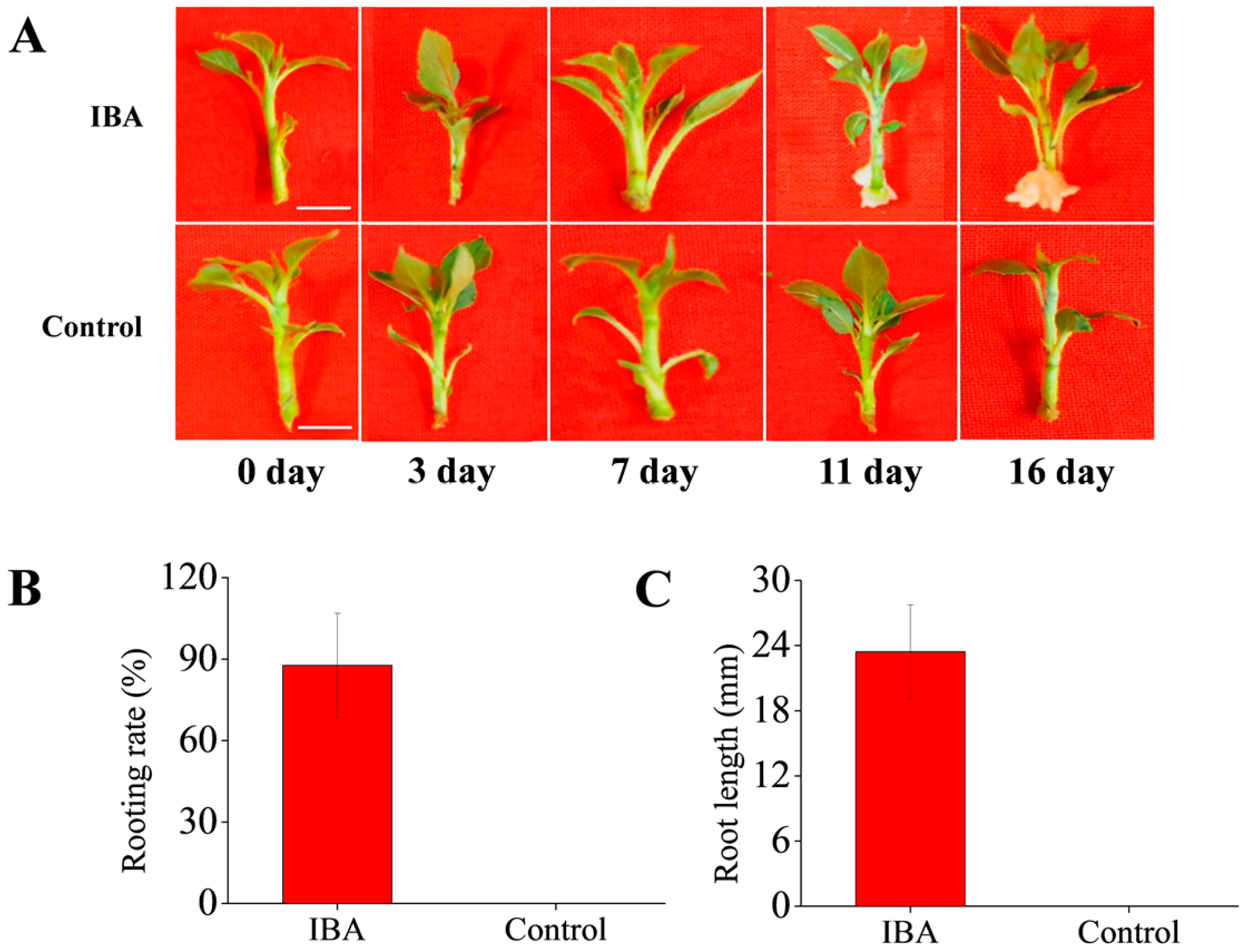
2.1. Effects of Exogenous IBA on AR Formation
2.2. Hormonal Changes in ‘T337’ Basal Stem Cuttings during AR Formation
2.3. Potential Protein Identification
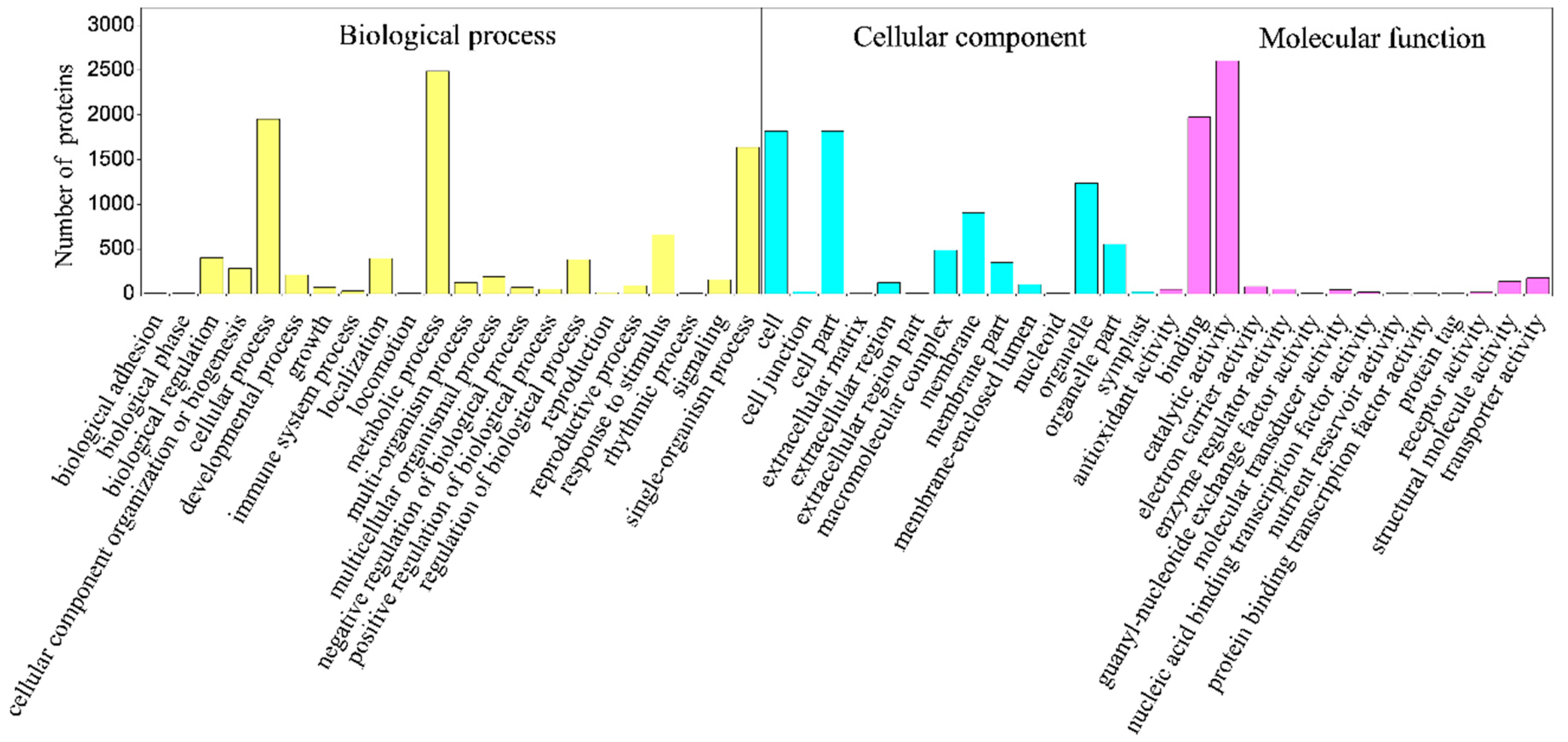
2.4. Classification of Identified Proteins
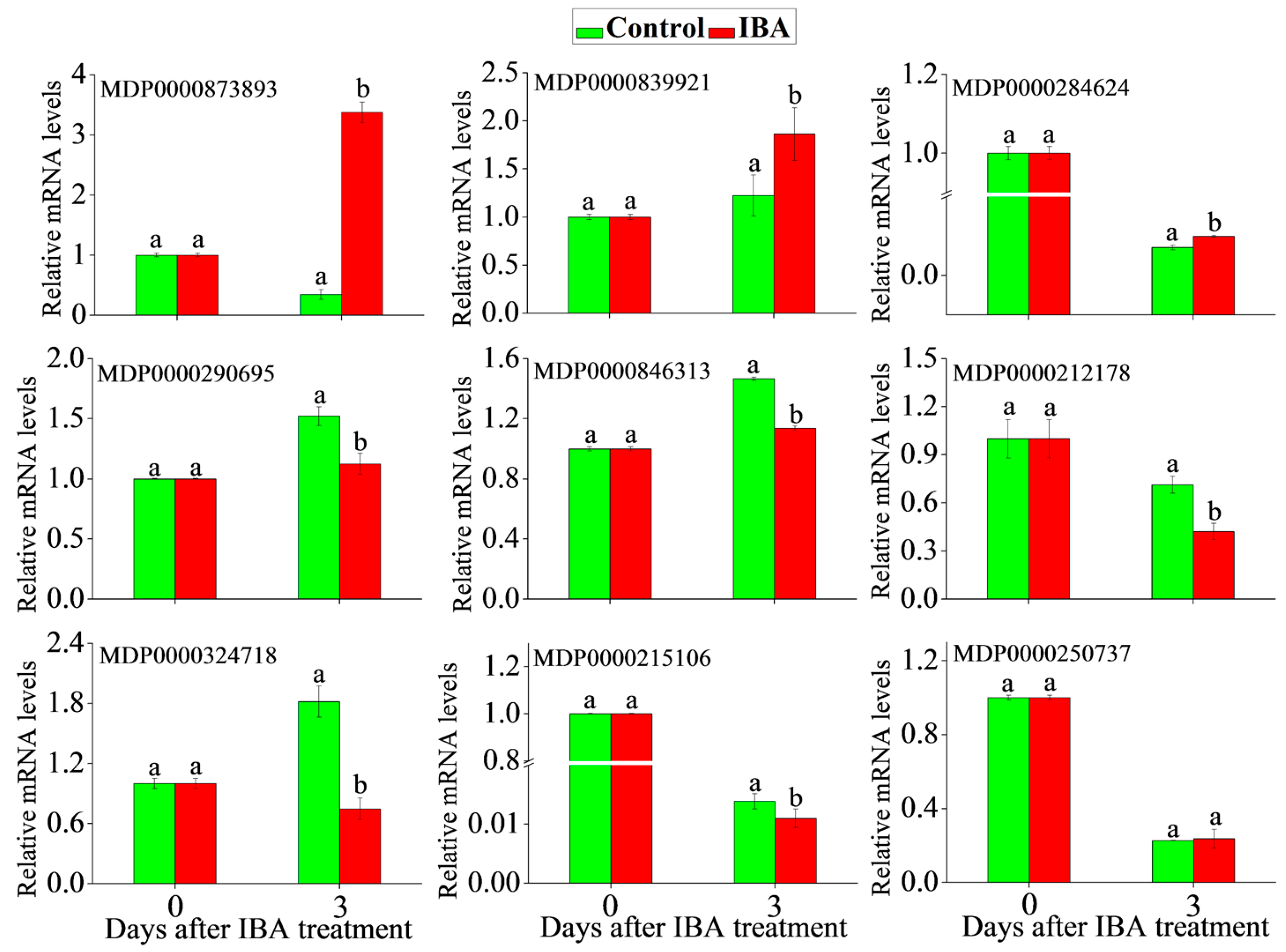
2.5. RT-qPCR Analysis
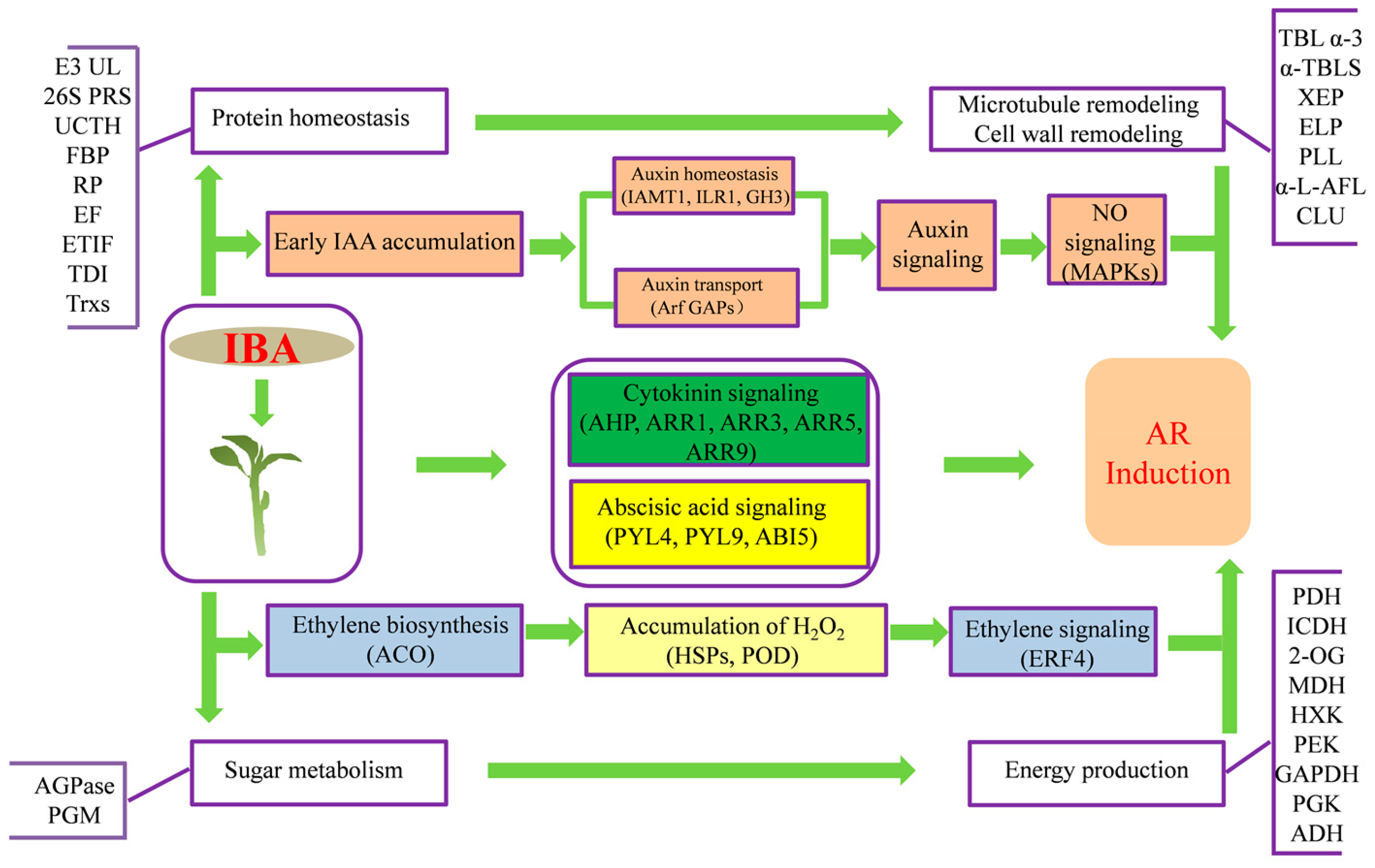
3. Discussion
3.1. IBA May Regulate Phytohormone Signaling to Contribute to AR Induction
3.2. Carbohydrate Metabolism and Energy Production may be Enhanced in Response to IBA
3.3. ROS and NO Signaling May Be Activated in Response to IBA
3.4. IBA Treatment May Change Protein Homeostasis, Especially Ubiquitinylation-Based Protein Degradation
3.5. The Effect of IBA on Microtubules and Cell Wall Properties
4. Materials and Methods
4.1. Plant Material
4.2. Measurement of Hormone Contents
4.3. Protein Extraction
4.4. iTRAQ Labeling and Strong Cation Exchange
4.5. LC-ESI-MS/MS Analysis
4.6. Protein Identification and Functional Annotation
4.7. Reverse Transcription-Quantitative PCR (RT-qPCR)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| iTRAQ | Isobaric tags for relative and absolute quantification |
| AR | Adventitious root |
| LR | Lateral root |
| IBA | Indole-3-butyric acid |
| IAA | Indole-3-acetic acid |
| CK | Cytokinin |
| ABA | Abscisic acid |
| GA | Gibberellin |
| JA | Jasmonic acid |
| BR | Brassinolide |
| HPLC | High Performance Liquid Chromatography |
| CHAPS | 3-[(3-Cholamidopropyl) dimethylammonio]propanesulfo |
| PMSF | Phenylmethylsulfonyl fluoride |
| EDTA | Ethylene Diamine Tetraacetic Acid |
| MS | Murashige and Skoog salts |
| TCA | Trichloroacetic Acid |
| DTT | dl-Dithiothreitol |
| IAM | Iodoacetamide |
| TEAB | Tetraethyl-ammonium bromide |
| ACN | Acetonitrile |
| BSA | Bull Serum Albumin |
| FA | Formic Acid |
| FDR | False Discovery Rate |
| NP40 | Nonidet P-40 |
References
- Li, S.W.; Xue, L.G.; Xu, S.J.; Feng, H.Y.; An, L.Z. Mediators, genes and signaling in adventitious rooting. Bot. Rev. 2009, 75, 230–247. [Google Scholar] [CrossRef]
- Abu-Abied, M.; Mordehaev, I.; Kumar, G.B.S.; Ophir, R.; Wasteneys, G.O.; Sadot, E. Analysis of microtubule-associated-proteins during IBA-mediated adventitious root induction reveals katanin dependent and independent alterations of expression patterns. PLoS ONE 2015, 10, e0143828. [Google Scholar] [CrossRef] [PubMed]
- Kevers, C.; Hausman, J.F.; Faivre-Rampant, O.; Evers, D.; Gaspar, T. Hormonal control of adventitious rooting: Progress and questions. J. Appl. Bot. Angew. Bot. 1997, 71, 71–79. [Google Scholar]
- De Klerk, G.J.; Van der Krieken, W.; De Jong, J.C. Review—The formation of adventitious roots: New concepts, new possibilities. In Vitro Cell. Dev. Biol. Plant 1999, 35, 189–199. [Google Scholar] [CrossRef]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Falasca, G.; Altamura, M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 2013, 112, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Bussell, J.D.; Pacurar, D.I.; Schwambach, J.; Pacurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of auxin response factor transcripts and microrna abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Armengot, L.; Marques-Bueno, M.M.; Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 2016, 67, 4015–4037. [Google Scholar] [CrossRef] [PubMed]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized induction of the ATP-binding cassette b19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Petrasek, J. Why plants need more than one type of auxin. Plant Sci. Int. J. Exp. Plant Biol. 2011, 180, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Heloir, M.C.; Kevers, C.; Hausman, J.F.; Gaspar, T. Changes in the concentrations of auxins and polyamines during rooting of in vitro-propagated walnut shoots. Tree Physiol. 1996, 16, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Muller, J.; Vertocnik, A.; Town, C.D. Analysis of indole-3-butyric acid-induced adventitious root formation on Arabidopsis stem segments. J. Exp. Bot. 2005, 56, 2095–2105. [Google Scholar] [CrossRef] [PubMed]
- Rout, G.R. Effect of auxins on adventitious root development from single node cuttings of camellia sinensis (L.) kuntze and associated biochemical changes. Plant Growth Regul. 2006, 48, 111–117. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.; Cheng, H.; Zhang, C.; Ma, C.; Zhang, L.; Gong, W.; Wu, L. Identification of genes involved in indole-3-butyric acid-induced adventitious root formation in nodal cuttings of camellia sinensis (L.) by suppression subtractive hybridization. Gene 2013, 514, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Robert, S.; Kleine-Vehn, J. Auxin: Simply complicated. J. Exp. Bot. 2013, 64, 2565–2577. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Carvajal, G.A.; Morse, A.M.; Dervinis, C.; Davis, J.M. The cytokinin type-b response regulator PTRR13 is a negative regulator of adventitious root development in populus. Plant Physiol. 2009, 150, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Veloccia, A.; Fattorini, L.; Della Rovere, F.; Sofo, A.; D’Angeli, S.; Betti, C.; Falasca, G.; Altamura, M.M. Ethylene and auxin interaction in the control of adventitious rooting in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 6445–6458. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.F.; Yang, S.Y.; Chen, K.T.; Hsing, Y.L.; Zeevaart, J.A.D.; Chen, L.J.; Yu, S.M. A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Debi, B.R.; Taketa, S.; Ichii, M. Cytokinin inhibits lateral root initiation but stimulates lateral root elongation in rice (oryza sativa). J. Plant Physiol. 2005, 162, 507–515. [Google Scholar] [CrossRef] [PubMed]
- da Costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Zhang, D.; Lei, C.; Chen, H.F.; Xing, L.B.; Ma, J.J.; Zhao, C.P.; Han, M.Y. Proteome analyses using iTRAQ labeling reveal critical mechanisms in alternate bearing malus prunifolia. J. Proteome Res. 2016, 15, 3602–3616. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.S.; Dong, S.R.; Zhang, H.Y.; Wang, S.Y. Putative model based on iTRAQ proteomics for Spirulina morphogenesis mechanisms. J. Proteom. 2017, 171, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Chang, R.; Lv, B.X.; Li, B.S. Quantitative proteomics analysis by iTRAQ revealed underlying changes in thermotolerance of Arthrospira platensis. J. Proteom. 2017, 165, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.W.; Ji, J.; Ma, X.T.; Xu, Q.; Qi, X.H.; Chen, X.H. Comparative proteomic analysis provides insight into the key proteins involved in cucumber (cucumis sativus L.) adventitious root emergence under waterlogging stress. Front. Plant Sci. 2016, 7, 1515. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Negroni, L.; Balliau, T.; Corti, H.; Jacquemot, M.P.; Davanture, M.; Sandberg, G.; Zivy, M.; Bellini, C. Proteomic analysis of different mutant genotypes of Arabidopsis led to the identification of 11 proteins correlating with adventitious root development. Plant Physiol. 2006, 140, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Welander, M. In vitro rooting of the apple rootstock m 26 in adult and juvenile growth phases and acclimatization of the plantlets. Physiol. Plant. 1983, 58, 231–238. [Google Scholar] [CrossRef]
- Jásik, J.; De Klerk, G.-J. Anatomical and ultrastructural examination of adventitious root formation in stem slices of apple. Biol. Plant. 1997, 39, 79–90. [Google Scholar] [CrossRef]
- Atkinson, J.A.; Rasmussen, A.; Traini, R.; Voss, U.; Sturrock, C.; Mooney, S.J.; Wells, D.M.; Bennett, M.J. Branching out in roots: Uncovering form, function, and regulation. Plant Physiol. 2014, 166, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Stepaniants, S.B.; Mao, M.; Weng, L.; Feetham, M.C.; Doyle, M.J.; Yi, E.C.; Dai, H.Y.; Thorsson, V.; Eng, J.; et al. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteom. 2004, 3, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Garrido, G.; Guerrero, J.R.; Cano, E.A.; Acosta, M.; Sanchez-Bravo, J. Origin and basipetal transport of the IAA responsible for rooting of carnation cuttings. Physiol. Plant. 2002, 114, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Bellamine, J.; Penel, C.; Greppin, H.; Gaspar, T. Confirmation of the role of auxin and calcium in the late phases of adventitious root formation. Plant Growth Regul. 1998, 26, 191–194. [Google Scholar] [CrossRef]
- Carranza, A.P.S.; Singh, A.; Steinberger, K.; Panigrahi, K.; Palme, K.; Dovzhenko, A.; Dal Bosco, C. Hydrolases of the ILR1-like family of Arabidopsis thaliana modulate auxin response by regulating auxin homeostasis in the endoplasmic reticulum. Sci. Rep. 2016, 6, 24212. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, R.; Ma, C.J.; Vlot, A.C.; Klessig, D.F.; Pichersky, E. Inactive methyl indole-3-acetic acid ester can be hydrolyzed and activated by several esterases belonging to the atmes esterase family of Arabidopsis. Plant Physiol. 2008, 147, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Anders, N.; Wolters, H.; Keicher, J.; Kornberger, W.; Muller, P.; Delbarre, A.; Ueda, T.; Nakano, A.; Jurgens, G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 2003, 112, 219–230. [Google Scholar] [CrossRef]
- Min, M.K.; Kim, S.J.; Miao, Y.; Shin, J.; Jiang, L.; Hwang, I. Overexpression of Arabidopsis AGD7 causes relocation of GOLGI-localized proteins to the endoplasmic reticulum and inhibits protein trafficking in plant cells. Plant Physiol. 2007, 143, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- DeKlerk, G.J.; ArnholdtSchmitt, B.; Lieberei, R.; Neumann, K.H. Regeneration of roots, shoots and embryos: Physiological, biochemical and molecular aspects. Biol. Plant. 1997, 39, 53–66. [Google Scholar] [CrossRef]
- Hwang, I.; Sheen, J.; Muller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Uhlir, N.J.; Reed, J.W. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 2002, 14, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Voesenek, L.A.; Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 2015, 206, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tian, L.N.; Latoszek-Green, M.; Brown, D.; Wu, K.Q. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 2005, 58, 585–596. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Jiang, L.; Xin, Q.; Liu, Y.; Tan, J.X.; Chen, Z.Z. Structural basis and functions of abscisic acid receptors PYLS. Front. Plant Sci. 2015, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Brocard, I.M.; Lynch, T.J.; Finkelstein, R.R. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002, 129, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Hisano, H.; Hojo, Y.; Matsuura, T.; Ikeda, Y.; Mori, I.C.; Senthil-Kumar, M. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals aba and JA as major regulators. Sci. Rep. 2017, 7, 4017. [Google Scholar] [CrossRef] [PubMed]
- Mauriat, M.; Petterle, A.; Bellini, C.; Moritz, T. Gibberellins inhibit adventitious rooting in hybrid aspen and Arabidopsis by affecting auxin transport. Plant J. Cell Mol. Biol. 2014, 78, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Busov, V.; Meilan, R.; Pearce, D.W.; Rood, S.B.; Ma, C.P.; Tschaplinski, T.J.; Strauss, S.H. Transgenic modification of GAI or RGL1 causes dwarfing and alters gibberellins, root growth, and metabolite profiles in populus. Planta 2006, 224, 288–299. [Google Scholar] [CrossRef] [PubMed]
- Sanders, P.M.; Lee, P.Y.; Biesgen, C.; Boone, J.D.; Beals, T.P.; Weiler, E.W.; Goldberg, R.B. The Arabidopsis delayed dehiscence1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 2000, 12, 1041–1061. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mongelard, G.; Flokova, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Han, C.Y.; Peng, W.; Peng, Z.H.; Xiong, X.Y.; Zhu, Q.; Gao, B.D.; Xie, D.X.; Ren, C.M. Brassinosteroid negatively regulates jasmonate inhibition of root growth in Arabidopsis. Plant Signal. Behav. 2010, 5, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Zerche, S.; Kadner, R. Nitrogen- and storage-affected carbohydrate partitioning in high-light-adapted pelargonium cuttings in relation to survival and adventitious root formation under low light. Ann. Bot. 2004, 94, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Klopotek, Y.; Haensch, K.T.; Hause, B.; Hajirezaei, M.R.; Druege, U. Dark exposure of petunia cuttings strongly improves adventitious root formation and enhances carbohydrate availability during rooting in the light. J. Plant Physiol. 2010, 167, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Gao, X.; Geng, J.; Li, Q.; Li, L.; Lv, Q.; Li, X. Identification of key proteins and networks related to grain development in wheat (triticum aestivum L.) by comparative transcription and proteomic analysis of allelic variants in TAGW2-6a. Front. Plant Sci. 2016, 7, 922. [Google Scholar] [CrossRef] [PubMed]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef] [PubMed]
- Holness, M.J.; Sugden, M.C. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem. Soc. Trans. 2003, 31, 1143–1151. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Martin, M.A.; Lopez-Lozano, A.; Diez, J.; Gomez-Baena, G.; Rangel-Zuniga, O.A.; Garcia-Fernandez, J.M. Physiological regulation of isocitrate dehydrogenase and the role of 2-oxoglutarate in prochlorococcus sp. Strain PCC 9511. PLoS ONE 2014, 9, e103380. [Google Scholar] [CrossRef] [PubMed]
- Wegener, K.M.; Singh, A.K.; Jacobs, J.M.; Elvitigala, T.; Welsh, E.A.; Keren, N.; Gritsenko, M.A.; Ghosh, B.K.; Camp, D.G., II; Smith, R.D.; et al. Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol. Cell. Proteom. MCP 2010, 9, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C. The organization and regulation of plant glycolysis. Annu. Rev. Plant Phys. 1996, 47, 185–214. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Kovalev, A.; Gorb, S.N.; Sauter, M. Emerging roots alter epidermal cell fate through mechanical and reactive oxygen species signaling. Plant Cell 2012, 24, 3296–3306. [Google Scholar] [CrossRef] [PubMed]
- Pucciariello, C.; Parlanti, S.; Banti, V.; Novi, G.; Perata, P. Reactive oxygen species-driven transcription in Arabidopsis under oxygen deprivation. Plant Physiol. 2012, 159, 184–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide involvement in formation and development of adventitious roots in cucumber. Plant Growth Regul. 2007, 52, 173–180. [Google Scholar] [CrossRef]
- Li, S.-W.; Xue, L.; Xu, S.; Feng, H.; An, L. Hydrogen peroxide acts as a signal molecule in the adventitious root formation of mung bean seedlings. Environ. Exp. Bot. 2009, 65, 63–71. [Google Scholar] [CrossRef]
- Lanteri, L.; Pagnussat, G.; Laxalt, A.; Lamattina, L. Nitric oxide is downstream of auxin and is required for inducing adventitious root formation in herbaceous and woody plants. In Adventitious Root Formation of Forest Trees and Horticultural Plants—From Genes to Applications; Research Signpost: Thiruvananthapuram, India, 2009; pp. 222–245. [Google Scholar]
- Yadav, S.; David, A.; Baluska, F.; Bhatla, S.C. Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signal. Behav. 2013, 8, e23196. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Yu, J.; Liao, W.; Yu, J.; Zhang, M.; Dawuda, M.M. Calcium and calmodulin are involved in Nitric Oxide-induced adventitious rooting of cucumber under simulated osmotic stress. Front. Plant Sci. 2017, 8, 1684. [Google Scholar] [CrossRef] [PubMed]
- Pagnussat, G.C.; Lanteri, M.L.; Lombardo, M.C.; Lamattina, L. Nitric oxide mediates the indole acetic acid induction activation of a mitogen-activated protein kinase cascade involved in adventitious root development. Plant Physiol. 2004, 135, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Urade, R. Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS J. 2007, 274, 1152–1171. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Boston, R.S. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 2008, 9, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. The ubiquitin-26s proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Vierstra, R.D. Proteolysis in plants: Mechanisms and functions. Plant Mol. Biol. 1996, 32, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Santner, A.; Estelle, M. The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. Cell Mol. Biol. 2010, 61, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Maraschin Fdos, S.; Memelink, J.; Offringa, R. Auxin-induced, SCF(TIR1)-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J. Cell Mol. Biol. 2009, 59, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Harberd, N.P.; Belfield, E.; Yasumura, Y. The angiosperm gibberellin-gid1-della growth regulatory mechanism: How an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 2009, 21, 1328–1339. [Google Scholar] [CrossRef] [PubMed]
- Thines, B.; Katsir, L.; Melotto, M.; Niu, Y.; Mandaokar, A.; Liu, G.; Nomura, K.; He, S.Y.; Howe, G.A.; Browse, J. Jaz repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Villacorta-Martin, C.; Sanchez-Garcia, A.B.; Villanova, J.; Cano, A.; van de Rhee, M.; de Haan, J.; Acosta, M.; Passarinho, P.; Perez-Perez, J.M. Gene expression profiling during adventitious root formation in carnation stem cuttings. BMC Genom. 2015, 16, 789. [Google Scholar] [CrossRef] [PubMed]
- Abu-Abied, M.; Rogovoy Stelmakh, O.; Mordehaev, I.; Grumberg, M.; Elbaum, R.; Wasteneys, G.O.; Sadot, E. Dissecting the contribution of microtubule behaviour in adventitious root induction. J. Exp. Bot. 2015, 66, 2813–2824. [Google Scholar] [CrossRef] [PubMed]
- Brinker, M.; van Zyl, L.; Liu, W.; Craig, D.; Sederoff, R.R.; Clapham, D.H.; von Arnold, S. Microarray analyses of gene expression during adventitious root development in pinus contorta. Plant Physiol. 2004, 135, 1526–1539. [Google Scholar] [CrossRef] [PubMed]
- Dobrev, P.I.; Vankova, R. Quantification of abscisic acid, cytokinin, and auxin content in salt-stressed plant tissues. Methods Mol. Biol. 2012, 913, 251–261. [Google Scholar] [PubMed]
- Ding, J.; Mao, L.J.; Wang, S.T.; Yuan, B.F.; Feng, Y.Q. Determination of endogenous brassinosteroids in plant tissues using solid-phase extraction with double layered cartridge followed by high-performance liquid chromatography-tandem mass spectrometry. Phytochem. Anal. PCA 2013, 24, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Li, Y.; Chen, Z.; Liu, Z.; Ren, Q.; Yang, J.; Zhu, X.; Guan, G.; Liu, A.; Luo, J.; et al. Comparative proteomic and bioinformatic analysis of theileria luwenshuni and theileria uilenbergi. Exp. Parasitol. 2016, 166, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Liang, J.R.; Chen, D.D.; Xu, B.; Zhuo, W.H.; Gao, Y.H.; Chen, C.P.; Bowler, C.; Zhang, W. iTRAQ-based proteomic analysis of the metabolism mechanism associated with silicon response in the marine diatom thalassiosira pseudonana. J. Proteome Res. 2014, 13, 720–734. [Google Scholar] [CrossRef] [PubMed]
- Wen, B.; Zhou, R.; Feng, Q.; Wang, Q.; Wang, J.; Liu, S. Iquant: An automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 2014, 14, 2280–2285. [Google Scholar] [CrossRef] [PubMed]
- Brosch, M.; Yu, L.; Hubbard, T.; Choudhary, J. Accurate and sensitive peptide identification with mascot percolator. J. Proteome Res. 2009, 8, 3176–3181. [Google Scholar] [CrossRef] [PubMed]
- Savitski, M.M.; Wilhelm, M.; Hahne, H.; Kuster, B.; Bantscheff, M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteom. MCP 2015, 14, 2394–2404. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. Kegg for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


| Pathway | DEPs (2868) a | All Proteins (6476) b | Pathway ID | p-Value c |
|---|---|---|---|---|
| Metabolic pathways | 873 (30.44%) | 1845 (28.49%) | ko01100 | 0.001077 |
| Biosynthesis of secondary metabolites | 505 (17.61%) | 1067 (16.48%) | ko01110 | 0.01567 |
| Starch and sucrose metabolism | 108 (3.77%) | 216 (3.34%) | ko00500 | 0.049829 |
| Ribosome | 135 (4.71%) | 237 (3.66%) | ko03010 | 0.0000442 |
| Phenylpropanoid biosynthesis | 91 (3.17%) | 140 (2.16%) | ko00940 | 0.000000492 |
| Pentose and glucuronate interconversions | 77 (2.68%) | 142 (2.19%) | ko00040 | 0.010236 |
| Peroxisome | 68 (2.37%) | 121 (1.87%) | ko04146 | 0.005217 |
| Carbon fixation in photosynthetic organisms | 56 (1.95%) | 104 (1.61%) | ko00710 | 0.030497 |
| Glycine, serine and threonine metabolism | 51 (1.78%) | 87 (1.34%) | ko00260 | 0.00477 |
| Glutathione metabolism | 50 (1.74%) | 85 (1.31%) | ko00480 | 0.004702 |
| Glyoxylate and dicarboxylate metabolism | 40 (1.39%) | 68 (1.05%) | ko00630 | 0.010849 |
| Cyanoamino acid metabolism | 40 (1.39%) | 70 (1.08%) | ko00460 | 0.020206 |
| beta-Alanine metabolism | 34 (1.19%) | 52 (0.8%) | ko00410 | 0.001692 |
| Phenylalanine metabolism | 34 (1.19%) | 53 (0.82%) | ko00360 | 0.00273 |
| Flavonoid biosynthesis | 34 (1.19%) | 54 (0.83%) | ko00941 | 0.004267 |
| Other glycan degradation | 34 (1.19%) | 57 (0.88%) | ko00511 | 0.013751 |
| Fatty acid degradation | 30 (1.05%) | 53 (0.82%) | ko00071 | 0.047544 |
| alpha-Linolenic acid metabolism | 29 (1.01%) | 49 (0.76%) | ko00592 | 0.025178 |
| Terpenoid backbone biosynthesis | 29 (1.01%) | 50 (0.77%) | ko00900 | 0.035041 |
| Photosynthesis | 27 (0.94%) | 42 (0.65%) | ko00195 | 0.00701 |
| Ubiquinone and other terpenoid-quinone biosynthesis | 25 (0.87%) | 39 (0.6%) | ko00130 | 0.009848 |
| Sphingolipid metabolism | 25 (0.87%) | 42 (0.65%) | ko00600 | 0.033381 |
| Tropane, piperidine and pyridine alkaloid biosynthesis | 21 (0.73%) | 31 (0.48%) | ko00960 | 0.007088 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 20 (0.7%) | 29 (0.45%) | ko00945 | 0.006307 |
| Limonene and pinene degradation | 17 (0.59%) | 20 (0.31%) | ko00903 | 0.000214 |
| Lysine degradation | 17 (0.59%) | 26 (0.4%) | ko00310 | 0.024492 |
| Photosynthesis—antenna proteins | 7 (0.24%) | 8 (0.12%) | ko00196 | 0.016319 |
| Phenylpropanoid biosynthesis | 91 (3.17%) | 140 (2.16%) | ko00940 | 0.000000492 |
| Accession No. a | Description | SwissProt Accession b | Identity (%) c | %COV (95) d | Unique Peptides | Ratio e |
|---|---|---|---|---|---|---|
| Phytohormone signaling | ||||||
| MDP0000873893 | Indole-3-acetic acid-amido synthetase | O82333 | 80.27 | 5.7 | 1 | 2.03 |
| MDP0000121609 | Indole-3-acetic acid-amido synthetase | O82333 | 80.14 | 17 | 2 | 1.64 |
| MDP0000310711 | IAA-amino acid hydrolase ILR1 | P54968 | 54.53 | 4.2 | 2 | 1.74 |
| MDP0000663451 | IAA-amino acid hydrolase ILR1 | P54968 | 62.83 | 10 | 3 | 1.52 |
| MDP0000290695 | Indole-3-acetate O-methyltransferase 1 | Q9FLN8 | 73.58 | 1.8 | 1 | 0.43 |
| MDP0000140463 | ADP-ribosylation factor gtpase-activating protein | O80925 | 70.28 | 6.8 | 3 | 0.67 |
| MDP0000250432 | ADP-ribosylation factor gtpase-activating protein | O80925 | 54.84 | 1.4 | 1 | 0.58 |
| MDP0000288128 | ADP-ribosylation factor gtpase-activating protein | O80925 | 62.68 | 1.3 | 1 | 0.37 |
| MDP0000186518 | Histidine-containing phosphotransfer protein 3 | Q9SAZ5 | 56.1 | 6.1 | 1 | 0.32 |
| MDP0000509768 | Arabidopsis response regulator ARR9 | O80366 | 59.18 | 5.7 | 1 | 0.45 |
| MDP0000212178 | Arabidopsis response regulator ARR5 | Q9ZWS6 | 78.01 | 4 | 1 | 0.53 |
| MDP0000250737 | Arabidopsis response regulator ARR3 | Q9ZWS9 | 76.61 | 5.9 | 2 | 0.29 |
| MDP0000846313 | Arabidopsis response regulator ARR1 | Q940D0 | 63.64 | 5.7 | 1 | 0.65 |
| MDP0000175691 | 1-aminocyclopropane-1-carboxylate oxidase 1 | Q9LSW7 | 29.34 | 6 | 1 | 1.57 |
| MDP0000663852 | 1-aminocyclopropane-1-carboxylate oxidase 1 | Q0WPW4 | 46.59 | 4.8 | 1 | 1.6 |
| MDP0000839921 | 1-aminocyclopropane-1-carboxylate oxidase 1 | Q84MB3 | 47.74 | 7 | 2 | 1.81 |
| MDP0000195885 | 1-aminocyclopropane-1-carboxylate oxidase 1 | Q00985 | 100 | 7.6 | 2 | 0.29 |
| MDP0000324718 | Ethylene-responsive transcription factor 4 | O80340 | 74.71 | 5.6 | 1 | 0.57 |
| MDP0000284624 | Abscisic acid receptor PYL9 | Q84MC7 | 81.29 | 6 | 1 | 1.56 |
| MDP0000215106 | Abscisic acid-insensitive 5-like protein 2 | Q9LES3 | 52.22 | 3.2 | 1 | 0.51 |
| MDP0000228470 | Abscisic acid receptor PYL4 | O80920 | 80.12 | 4.6 | 1 | 0.58 |
| Carbohydrate metabolism and energy production | ||||||
| MDP0000866748 | Phosphoglucomutase | Q9SCY0 | 84.31 | 14.9 | 7 | 1.79 |
| MDP0000256619 | ADP-glucose pyrophosphorylase 1 | P52417 | 88.72 | 16.7 | 1 | 2.44 |
| MDP0000298815 | α-amylase | P17859 | 72.8 | 2.2 | 2 | 2.2 |
| MDP0000657082 | α-mannosidase | P34098 | 61.35 | 9.1 | 2 | 1.84 |
| MDP0000095637 | Granule-bound starch synthase 1 | O82627 | 74.56 | 21.2 | 9 | 2.69 |
| MDP0000133306 | d-sorbitol-6-phosphate dehydrogenase | P28475 | 84.52 | 11.9 | 1 | 2.62 |
| MDP0000661960 | Xyloglucan endotransglucosylase/hydrolase protein 6 | Q8LF99 | 78.36 | 14.4 | 2 | 2.7 |
| MDP0000129346 | α-1,4 glucan phosphorylase L isozyme | P53536 | 81.22 | 21.4 | 15 | 1.63 |
| MDP0000320017 | Xyloglucan endotransglucosylase/hydrolase protein | Q38910 | 75.76 | 2.8 | 1 | 1.5 |
| MDP0000296747 | α-glucosidase | Q9F234 | 45.19 | 8.7 | 5 | 1.5 |
| MDP0000177786 | 1,4-α-glucan-branching enzyme | P30924 | 73.7 | 10.7 | 7 | 1.5 |
| MDP0000202465 | β-galactosidase | Q9FN08 | 60.98 | 19 | 13 | 0.19 |
| MDP0000290090 | β-galactosidase 6 | Q10NX8 | 42.16 | 4.8 | 1 | 0.47 |
| MDP0000295518 | α-l-fucosidase 1 | Q8GW72 | 71.43 | 10.7 | 4 | 1.67 |
| MDP0000863563 | β-galactosidase 9 | Q9SCV3 | 77.39 | 16.6 | 5 | 1.71 |
| MDP0000237069 | α-galactosidase | Q9FXT4 | 25.52 | 8.4 | 2 | 1.71 |
| MDP0000823956 | Hexokinase | Q9SEK2 | 78.16 | 13.7 | 3 | 1.65 |
| MDP0000294262 | 6-phosphofructokinase | Q41141 | 82.18 | 8.1 | 1 | 0.4 |
| MDP0000254412 | 6-phosphofructokinase | Q8VYN6 | 79.88 | 5.5 | 1 | 1.55 |
| MDP0000835914 | Glyceraldehyde-3-phosphate dehydrogenase | P12859 | 88.47 | 16.2 | 5 | 1.62 |
| MDP0000527995 | Glyceraldehyde-3-phosphate dehydrogenase | P12858 | 88.15 | 24.8 | 6 | 1.69 |
| MDP0000543856 | Glyceraldehyde-3-phosphate dehydrogenase | Q8S0G4 | 79.87 | 6.5 | 1 | 1.84 |
| MDP0000174843 | 3-phosphoglycerate kinase | Q42961 | 88.13 | 22.2 | 1 | 1.63 |
| MDP0000212948 | 3-phosphoglycerate kinase | Q42961 | 84.14 | 23.8 | 1 | 0.57 |
| MDP0000211987 | Aldehyde dehydrogenase | Q9ZPB7 | 82.41 | 8.7 | 4 | 1.57 |
| MDP0000221713 | Aldehyde dehydrogenase | Q9SU63 | 82.05 | 18.4 | 7 | 2 |
| MDP0000236430 | Alcohol dehydrogenases | P42734 | 66.77 | 8.6 | 4 | 1.78 |
| MDP0000267169 | Fructose-1,6-bisphosphatase | P46283 | 75.31 | 4.9 | 1 | 2.07 |
| MDP0000273014 | Fructose-1,6-bisphosphatase | P46275 | 78.87 | 19.5 | 6 | 1.67 |
| MDP0000244771 | Fructose-1,6-bisphosphatase | P46283 | 83.72 | 5.8 | 3 | 1.57 |
| MDP0000277811 | Fructose-1,6-bisphosphatase | P46283 | 80.67 | 5 | 1 | 1.95 |
| MDP0000275261 | Probable fructokinase-2 | Q9LNE3 | 65.8 | 8.5 | 1 | 1.57 |
| MDP0000178814 | Pyruvate dehydrogenase complex | Q54M22 | 53.72 | 6.3 | 1 | 1.59 |
| MDP0000325085 | Isocitrate dehydrogenase | P50217 | 89.78 | 21.1 | 3 | 1.54 |
| MDP0000214399 | 2-oxoglutarate and Fe(II)-dependent oxygenase superfamily protein | Q39110 | 26.8 | 3.2 | 1 | 1.68 |
| MDP0000384593 | Malic enzyme | P12628 | 83.78 | 23.7 | 8 | 1.58 |
| MDP0000221561 | Malic enzyme | P51615 | 83.94 | 24.4 | 7 | 1.5 |
| MDP0000313179 | NADH: ubiquinone oxidoreductase 20 kd subunit | P42027 | 98.11 | 5.5 | 1 | 1.53 |
| MDP0000362465 | Cytochrome b-c1 complex subunit 9 | P46270 | 76.39 | 22.4 | 2 | 1.57 |
| MDP0000124616 | Cytochrome c oxidase subunit 5C-2 | Q9LZQ0 | 77.78 | 12.7 | 1 | 1.65 |
| MDP0000263444 | Cytochrome c oxidase subunit 6b-1 | Q9S7L9 | 84.06 | 4.5 | 1 | 0.41 |
| MDP0000385730 | ATP synthase gamma chain, chloroplastic | P29790 | 85.68 | 9.7 | 3 | 1.75 |
| MDP0000168167 | Atpases with chaperone activity, chloroplastic | Q9LF37 | 81.33 | 16.6 | 2 | 1.73 |
| MDP0000929055 | ATP synthase subunit alpha, mitochondrial | P05495 | 99.16 | 10.1 | 2 | 1.65 |
| MDP0000360515 | ATP synthase subunit beta, mitochondrial | P17614 | 93.16 | 38.9 | 2 | 1.59 |
| MDP0000785964 | ATP synthase subunit delta, mitochondrial | Q40089 | 92.11 | 13.9 | 1 | 0.41 |
| MDP0000624197 | ATP synthase subunit delta, mitochondrial | Q40089 | 78.5 | 15.4 | 3 | 0.39 |
| MDP0000416290 | Atpase 4, plasma membrane-type | Q9SU58 | 84.62 | 5.5 | 1 | 0.58 |
| ROS and NO signaling | ||||||
| MDP0000684170 | Heat shock 70 kda protein | Q9SKY8 | 69.76 | 3.3 | 1 | 1.54 |
| MDP0000697285 | Heat shock 70 kda protein | Q02028 | 79.23 | 12.4 | 1 | 1.64 |
| MDP0000319048 | Cationic peroxidase 1 | P22195 | 74.6 | 21.3 | 6 | 0.43 |
| MDP0000545323 | Peroxidase 42 | Q9SB81 | 83.55 | 8.4 | 1 | 0.47 |
| MDP0000243237 | Peroxidase 44 | Q93V93 | 56.11 | 33.5 | 7 | 0.35 |
| MDP0000208152 | Peroxidase 53 | Q42578 | 60.98 | 9.8 | 2 | 0.43 |
| MDP0000154541 | Peroxidase 53 | Q42578 | 63.5 | 13.2 | 1 | 0.66 |
| MDP0000192235 | Peroxidase 42 | Q9SB81 | 75.44 | 6.1 | 1 | 0.67 |
| MDP0000706473 | Peroxidase 53 | Q42578 | 63.61 | 17.4 | 2 | 0.67 |
| MDP0000283650 | Peroxidase 4 | A7NY33 | 62.46 | 7 | 2 | 0.62 |
| MDP0000136398 | Peroxidase 16 | Q96518 | 64.42 | 4.6 | 1 | 0.49 |
| MDP0000301828 | Peroxidase 53 | Q42578 | 79.22 | 12.7 | 2 | 0.49 |
| MDP0000209189 | Cationic peroxidase 1 | P22195 | 73.58 | 29 | 5 | 0.55 |
| MDP0000251955 | Mitogen-activated protein kinase | Q40353 | 86.38 | 5.9 | 2 | 1.51 |
| Protein homeostasis | ||||||
| MDP0000241084 | E3 ubiquitin-protein ligase RGLG1 | Q9SS90 | 71.97 | 6.7 | 1 | 1.51 |
| MDP0000317971 | E3 ubiquitin-protein ligase UPL2 | Q8H0T4 | 48.73 | 3.9 | 3 | 0.57 |
| MDP0000269081 | E3 ubiquitin-protein ligase RING1 | P0CH30 | 58.54 | 3.4 | 1 | 0.64 |
| MDP0000676693 | ATP-dependent 26S proteasome regulatory subunit | Q54DY9 | 34 | 22.7 | 4 | 2.03 |
| MDP0000322270 | ATP-dependent 26S proteasome regulatory subunit | Q7ZZ25 | 42.23 | 6.2 | 1 | 0.58 |
| MDP0000315993 | ATP-dependent Zn proteases | O80860 | 85.11 | 4.7 | 6 | 1.61 |
| MDP0000188831 | 20S proteasome, alpha and beta subunits | O82178 | 58.71 | 16.2 | 5 | 0.45 |
| MDP0000245541 | Ubiquitin carboxyl-terminal hydrolase 2 | Q8W4N3 | 51.36 | 0.9 | 1 | 0.45 |
| MDP0000283283 | Ubiquitin carboxyl-terminal hydrolase 24 | Q9FPS3 | 74.27 | 5.8 | 2 | 0.64 |
| MDP0000263256 | F-box protein | Q9FGY4 | 28.24 | 1.7 | 1 | 1.73 |
| MDP0000180936 | F-box protein | Q9FE83 | 54.18 | 4.9 | 1 | 2.38 |
| MDP0000141860 | 40S ribosomal protein S30 | P49689 | 98.39 | 16.1 | 1 | 0.38 |
| MDP0000265859 | 60S ribosomal protein L14-1 | Q9SIM4 | 87.72 | 14.7 | 2 | 0.53 |
| MDP0000865687 | 40S ribosomal protein S17-4 | Q9LZ17 | 83.69 | 17.9 | 1 | 0.39 |
| MDP0000544199 | 50S ribosomal protein L24 | A8LC45 | 49.51 | 10.9 | 1 | 0.39 |
| MDP0000283097 | 40S ribosomal protein S17-4 | Q9LZ17 | 79.43 | 7.3 | 1 | 0.45 |
| MDP0000266765 | 30S ribosomal protein S5 | P93014 | 68.2 | 9.5 | 3 | 0.64 |
| MDP0000772832 | 40S ribosomal protein S19-3 | Q9FNP8 | 78.06 | 25.2 | 2 | 0.58 |
| MDP0000417422 | 40S ribosomal protein S5 | O24111 | 90.24 | 18.7 | 1 | 0.59 |
| MDP0000169133 | Translation elongation factor | B7K735 | 62.8 | 2.6 | 2 | 0.44 |
| MDP0000903484 | Translation elongation factor EF-1 | P93447 | 62.83 | 21.5 | 2 | 0.44 |
| MDP0000800338 | Translation initiation factor IF-2 | P57997 | 70.64 | 5.5 | 1 | 0.62 |
| MDP0000142167 | Eukaryotic translation initiation factor 2 subunit | P55871 | 96.68 | 7.5 | 1 | 0.63 |
| MDP0000270113 | Eukaryotic translation initiation factor 3 subunit | Q40554 | 64.54 | 9.5 | 2 | 0.6 |
| MDP0000141898 | Eukaryotic translation initiation factor 3 subunit | Q38884 | 81.19 | 14.4 | 1 | 0.67 |
| MDP0000261642 | Tryptophanyl-tRNA ligase | Q5UPJ7 | 44.83 | 3.1 | 2 | 1.63 |
| MDP0000134153 | Histidine–tRNA ligase | P93422 | 33.14 | 2 | 1 | 1.61 |
| MDP0000770881 | Glycine–tRNA ligase 1 | O23627 | 58.43 | 8.5 | 1 | 1.56 |
| MDP0000207727 | Thiol-disulfide isomerase and thioredoxins | O80763 | 64.64 | 20.8 | 3 | 2.63 |
| MDP0000308890 | Thiol-disulfide isomerase and thioredoxins | O80763 | 62.36 | 15.5 | 3 | 2.39 |
| MDP0000297301 | Thiol-disulfide isomerase and thioredoxins | O80763 | 59.55 | 2.2 | 1 | 1.81 |
| MDP0000505556 | Cysteine protease | P25776 | 44.1 | 3.1 | 1 | 1.77 |
| Microtubules and Cell wall remolding | ||||||
| MDP0000812416 | Tubulin α-3 | P33627 | 96.05 | 18.3 | 2 | 1.58 |
| MDP0000282827 | α-tubulin suppressor | Q9P258 | 77.58 | 3.9 | 2 | 0.67 |
| MDP0000296747 | Xyloglucan endotransglucosylase/hydrolase | Q9F234 | 45.19 | 8.7 | 5 | 1.5 |
| MDP0000661960 | Xyloglucan endotransglucosylase/hydrolase | Q8LF99 | 78.36 | 14.4 | 2 | 2.7 |
| MDP0000640549 | Expansin-like protein | O23547 | 54.84 | 5.9 | 1 | 2.16 |
| MDP0000130769 | Pectin lyase-like superfamily protein | A7PZL3 | 52.81 | 12.5 | 4 | 1.53 |
| MDP0000248311 | Pectin lyase-like superfamily protein | P15922 | 26.05 | 5.2 | 3 | 1.72 |
| MDP0000943790 | Pectin lyase-like superfamily protein | Q949Z1 | 47.68 | 1.9 | 1 | 0.3 |
| MDP0000175757 | Pectin lyase-like superfamily protein | Q94AJ5 | 45.06 | 4.2 | 2 | 0.49 |
| MDP0000251956 | Pectin lyase-like superfamily protein | A7PZL3 | 80.32 | 17.4 | 4 | 0.49 |
| MDP0000753366 | Cellulase | P23548 | 24.41 | 4.8 | 1 | 2.32 |
| MDP0000055078 | α-l-arabinofuranosidase 1 | Q9SG80 | 70.21 | 16.8 | 5 | 1.57 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lei, C.; Fan, S.; Li, K.; Meng, Y.; Mao, J.; Han, M.; Zhao, C.; Bao, L.; Zhang, D. iTRAQ-Based Proteomic Analysis Reveals Potential Regulation Networks of IBA-Induced Adventitious Root Formation in Apple. Int. J. Mol. Sci. 2018, 19, 667. https://doi.org/10.3390/ijms19030667
Lei C, Fan S, Li K, Meng Y, Mao J, Han M, Zhao C, Bao L, Zhang D. iTRAQ-Based Proteomic Analysis Reveals Potential Regulation Networks of IBA-Induced Adventitious Root Formation in Apple. International Journal of Molecular Sciences. 2018; 19(3):667. https://doi.org/10.3390/ijms19030667
Chicago/Turabian StyleLei, Chao, Sheng Fan, Ke Li, Yuan Meng, Jiangping Mao, Mingyu Han, Caiping Zhao, Lu Bao, and Dong Zhang. 2018. "iTRAQ-Based Proteomic Analysis Reveals Potential Regulation Networks of IBA-Induced Adventitious Root Formation in Apple" International Journal of Molecular Sciences 19, no. 3: 667. https://doi.org/10.3390/ijms19030667





