Fine Physical Bin Mapping of the Powdery Mildew Resistance Gene Pm21 Based on Chromosomal Structural Variations in Wheat
Abstract
:1. Introduction
2. Results
2.1. Physical Mapping of Pm21
2.2. Comparative Genomics Analysis of 6VS FLb4–b5/b6
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Development of 6VS-Specific Markers
4.3. DNA Extraction and PCR
4.4. Physical Mapping of Pm21
4.5. Comparative Genomics Analysis of the Pm21 Locus
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chen, P.D.; Qi, L.L.; Zhou, B.; Zhang, S.Z.; Liu, D.J. Development and molecular cytogenetic analysis of wheat—Haynaldia villosa 6VS/6AL translocation lines specifying resistance to powdery mildew. Theor. Appl. Genet. 1995, 91, 1125–1128. [Google Scholar] [CrossRef] [PubMed]
- Cao, A.; Xing, L.; Wang, X.; Yang, X.; Wang, W.; Sun, Y.; Qian, C.; Ni, J.; Chen, Y.; Liu, D.; et al. Serine/threonine kinase gene Stpk-V, a key member of powdery mildew resistance gene Pm21, confers powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732. [Google Scholar] [CrossRef] [PubMed]
- Bie, T.; Zhao, R.; Zhu, S.; Chen, S.; Cen, B.; Zhang, B.; Gao, D.; Jiang, Z.; Chen, T.; Wang, L.; et al. Development and characterization of marker MBH1 simultaneously tagging genes Pm21 and PmV conferring resistance to powdery mildew in wheat. Mol. Breed. 2015, 35, 189. [Google Scholar] [CrossRef]
- Chen, S.; Chen, P.; Wang, X. Inducement of chromosome translocation with small alien segments by irradiating mature female gametes of the whole arm translocation line. Sci. China Ser. C 2008, 51, 346–352. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zhu, S.; Jiang, Z.; Ji, Y.; Wang, F.; Zhao, R.; Bie, T. Comparative mapping of powdery mildew resistance gene Pm21 and functional characterization of resistance-related genes in wheat. Theor. Appl. Genet. 2016, 129, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; You, C.; Hu, Y.; Chen, S.; Zhou, B.; Cao, A.; Wang, X. Radiation-induced translocations with reduced Haynaldia villosa chromatin at the Pm21 locus for powdery mildew resistance in wheat. Mol. Breed. 2013, 31, 477–484. [Google Scholar] [CrossRef]
- He, H.; Ji, Y.; Zhu, S.; Li, B.; Zhao, R.; Jiang, Z.; Bie, T. Genetic, physical and comparative mapping of the powdery mildew resistance gene Pm21 originating from Dasypyrum villosum. Front. Plant Sci. 2017, 8, 1914. [Google Scholar] [CrossRef] [PubMed]
- Bie, T.; Zhao, R.; Jiang, Z.; Gao, D.; Zhang, B.; He, H. Efficient marker-assisted screening of structural changes involving Haynaldia villosa chromosome 6V using a double-distal-marker strategy. Mol. Breed. 2015, 35, 34. [Google Scholar] [CrossRef]
- Qi, X.; Cui, F.; Li, Y.; Ding, A.; Li, J.; Chen, G.; Wang, H. Molecular tagging wheat powdery mildew resistance gene Pm21 by EST-SSR and STS markers. Mol. Plant Breed. 2010, 1, 22–26. [Google Scholar] [CrossRef]
- Song, W.; Xie, C.; Du, J.; Xie, H.; Liu, Q.; Ni, Z.; Yang, T.; Sun, Q.; Liu, Z. A “one-marker-for-two-genes” approach for efficient molecular discrimination of Pm12 and Pm21 conferring resistance to powdery mildew in wheat. Mol. Breed. 2009, 23, 357–363. [Google Scholar] [CrossRef]
- Qi, L.L.; Wang, S.L.; Chen, P.D.; Liu, D.J.; Gill, B.S. Identification and physical mapping of three Haynaldia villosa chromosome-6V deletion lines. Theor. Appl. Genet. 1998, 97, 1042–1046. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, M.; Zhang, Y.; Chu, C.; Lin, Z.; Xu, Q.; Ye, X.; Chen, X.; Zhang, X. Development and application of functional markers specific to powdery mildew resistance on chromosome arm 6VS from different origins of Haynaldia villosa. Acta Agron. Sin. 2012, 38, 1827–1832. [Google Scholar] [CrossRef]
- Cao, A.Z.; Wang, X.E.; Chen, Y.P.; Zou, X.W.; Chen, P.D. A sequence-specific PCR marker linked with Pm21 distinguishes chromosomes 6AS, 6BS, 6DS of Triticum aestivum and 6VS of Haynaldia villosa. Plant Breed. 2006, 125, 201–205. [Google Scholar] [CrossRef]
- Friebe, B.; Heun, M.; Tuleen, N.; Zeller, F.J.; Gill, B.S. Cytogenetically monitored transfer of powdery mildew resistance from rye into wheat. Crop Sci. 1994, 34, 621–625. [Google Scholar] [CrossRef]
- Jia, J.; Devos, K.M.; Chao, S.; Miller, T.E.; Reader, S.M.; Gale, M.D. RFLP-based maps of the homoeologous group-6 chromosomes of wheat and their application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theor. Appl. Genet. 1996, 92, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; D’Ovidio, R.; Tanzarella, O.A.; Ceoloni, C.; Porceddu, E. Identification of molecular markers linked to Pm13, an Aegilops longissima gene conferring resistance to powdery mildew in wheat. Theor. Appl. Genet. 1999, 98, 448–454. [Google Scholar] [CrossRef]
- He, H.; Zhu, S.; Sun, W.; Gao, D.; Bie, T. Efficient development of Haynaldia villosa chromosome 6VS-specific DNA markers using a CISP-IS strategy. Plant Breed. 2013, 132, 290–294. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed]


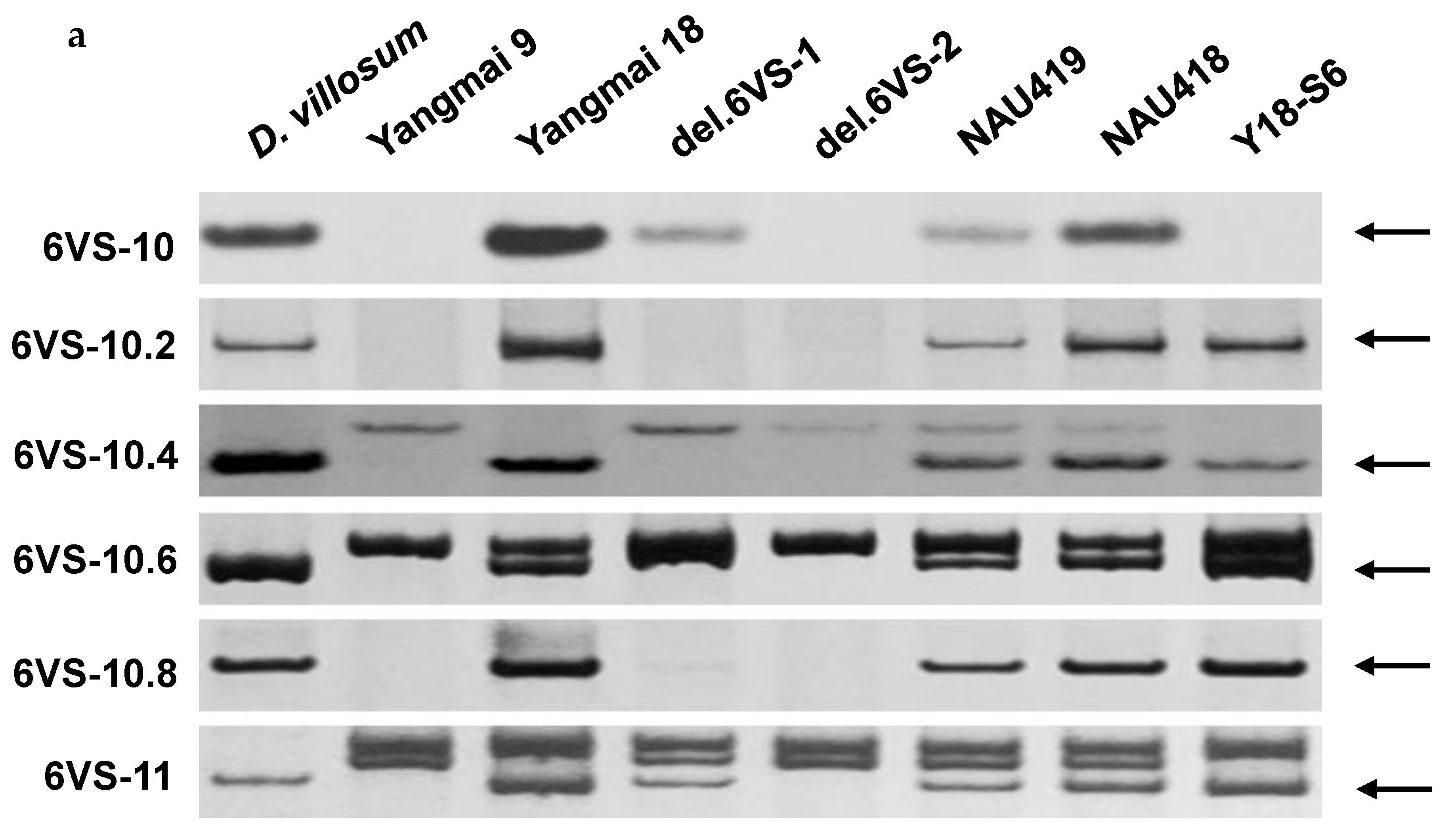
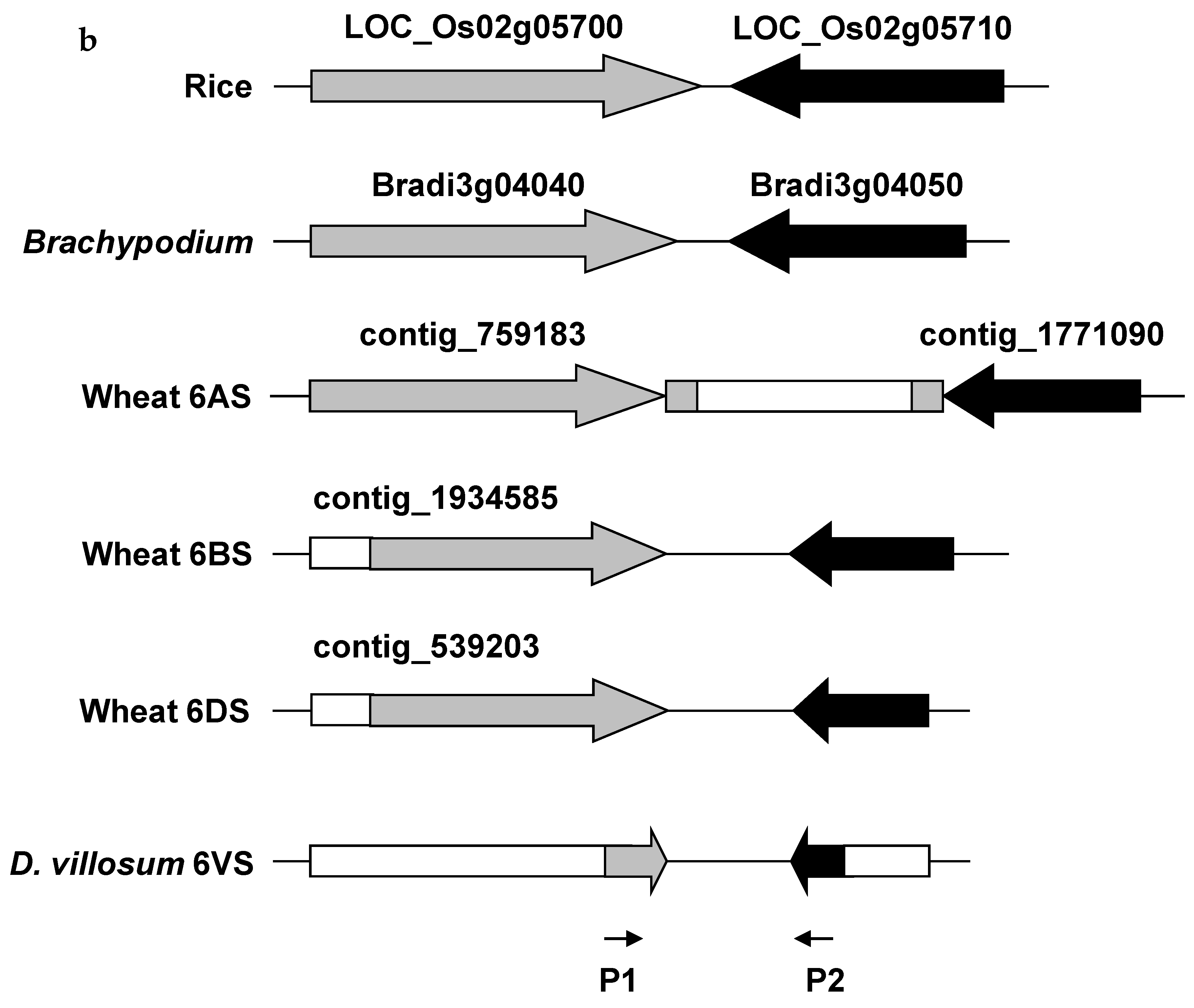
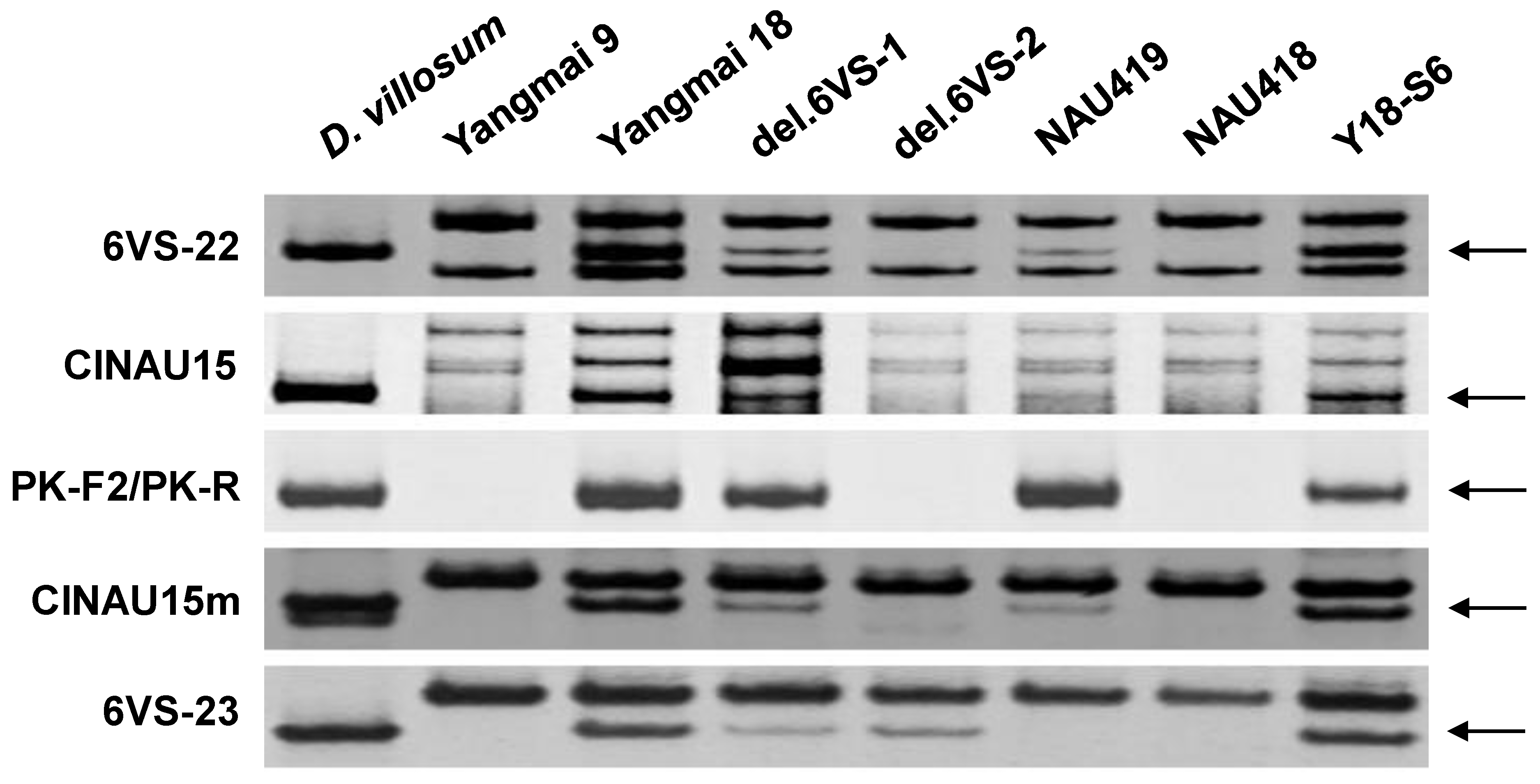
| Genetic Stock | Treatment | Chromosomal Breakpoint | Flanking Markers |
|---|---|---|---|
| del.6VS-1 | Irradiation a | b2 (FL0.58) | 6VS-03/6VS-04 |
| b6 | 6VS-10/6VS-10.2 | ||
| b7 | 6VS-10.8/6VS-11 | ||
| del.6VS-2 | Irradiation | b9 (FL0.45) | CINAU15m/6VS-23 |
| NAU418 | Irradiation | b4 | 6VS-08.6/6VS-08.8 |
| b8 | 6VS-13/6VS-13.5 | ||
| NAU419 | Irradiation | b1 | Xcinau188/6VS-01 |
| b10 (Close to b9) | CINAU15m/6VS-23 | ||
| Y18-S6 | EMS | b3 (Close to b2) | 6VS-03/6VS-04 |
| b5 (Close to b6) | 6VS-10/6VS-10.2 |
| Marker | Brachypodium | Rice | Wheat | Gene Annotation |
|---|---|---|---|---|
| 6VS-08.6 | Bradi3g03850 | 6AS_contigs_4399884 6BS_contigs_2953789 6DS_contigs_2117578 | Eukaryotic translation initiation factor | |
| 6VS-08.8 | Bradi3g03860 | LOC_Os02g05620 | 6AS_contigs_4431592 6BS_contigs_2953283 6DS_contigs_2092656 | Exocyst complex subunit EXO70-like protein |
| 6VS-09 | Bradi3g03870 | LOC_Os02g05630 | 6AS_contigs_4363243 6BS_contigs_2962596 6DS_contigs_2093935 | Serine/threonine protein phosphatase 2C |
| MBH1 | Bradi3g03874 a | Six contigs a | Disease resistance protein | |
| 6VS-09.4 | Bradi3g03878 a | Six contigs a | Disease resistance protein | |
| Bradi3g03882 a | Six contigs a | Disease resistance protein | ||
| Bradi3g03886 | Unknown protein | |||
| Bradi3g03890 | Unknown protein | |||
| Bradi3g03900 | Unknown protein | |||
| Bradi3g03910 | Cytochrome P450 | |||
| Bradi3g03920 | Unknown protein | |||
| Bradi3g03930 | Poly(A) polymerase | |||
| Bradi3g03935 a | Six contigs a | Disease resistance protein | ||
| Bradi3g03940 | 6AS_contigs_4428294 6BS_contigs_2926507 6DS_contigs_2114667 | Photosystem II protein J | ||
| Bradi3g03945 | 6AS_contigs_4428294 6BS_contigs_2926507 6DS_contigs_2114667 | Photosystem II cytochrome b559 alpha subunit | ||
| Bradi3g03950 | Unknown protein | |||
| Bradi3g03957 | 6AS_contigs_4431958 6BS_contigs_2953283 6DS_contigs_2081863 | Polyubiquitin | ||
| 6VS-10 | Bradi3g03970 | LOC_Os02g05640 | 6AS_contigs_4429249 b 6BS_contigs_1424756 b | Homeobox-leucine zipper protein HOX26 |
| 6DS_contigs_2087435 b | ||||
| LOC_Os02g05650 | Unknown protein | |||
| Bradi3g03980 | 6AS_contigs_2062052 | Unknown protein | ||
| 6DS_contigs_2110435 | ||||
| Bradi3g03990 | LOC_Os02g05660 | DEAD-box ATP-dependent RNA helicase | ||
| 6VS-10.2 | Bradi3g04000 | LOC_Os02g05661 | 6AS_contigs_4428740 6DS_contigs_2082952 | Unknown protein |
| Marker | Strategy | Primer Sequence (5′ → 3′) | Annealing Temperature (°C) | Size of Polymorphic Band (bp) |
|---|---|---|---|---|
| 6VS-08.6 | CISP-IS | F: GATCTCGTTTTAGTCCTGAGCCTGT | 60 | 604 |
| R: CCGAGTGAAGTCTGCTCATCCAGT | ||||
| 6VS-12.5 | CISP | F: CCGGCGACGCGCACTAC | 60 | 391 |
| R: GTACTTGTCCGCGAACTCGAAC | ||||
| 6VS-13.5 | CISP | F: TGCCTCATCACCCCAGTGAA | 60 | 287 |
| R: TGCAGTAGGTGGCATAGAGCAAG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, S.; Ji, Y.; Ji, J.; Bie, T.; Gao, A.; He, H. Fine Physical Bin Mapping of the Powdery Mildew Resistance Gene Pm21 Based on Chromosomal Structural Variations in Wheat. Int. J. Mol. Sci. 2018, 19, 643. https://doi.org/10.3390/ijms19020643
Zhu S, Ji Y, Ji J, Bie T, Gao A, He H. Fine Physical Bin Mapping of the Powdery Mildew Resistance Gene Pm21 Based on Chromosomal Structural Variations in Wheat. International Journal of Molecular Sciences. 2018; 19(2):643. https://doi.org/10.3390/ijms19020643
Chicago/Turabian StyleZhu, Shanying, Yaoyong Ji, Jian Ji, Tongde Bie, Anli Gao, and Huagang He. 2018. "Fine Physical Bin Mapping of the Powdery Mildew Resistance Gene Pm21 Based on Chromosomal Structural Variations in Wheat" International Journal of Molecular Sciences 19, no. 2: 643. https://doi.org/10.3390/ijms19020643





