Molecular Cloning and Characterization of a Wild Eggplant Solanum aculeatissimum NBS-LRR Gene, Involved in Plant Resistance to Meloidogyne incognita
Abstract
:1. Introduction
2. Results
2.1. Isolation and Characterization of SacMi
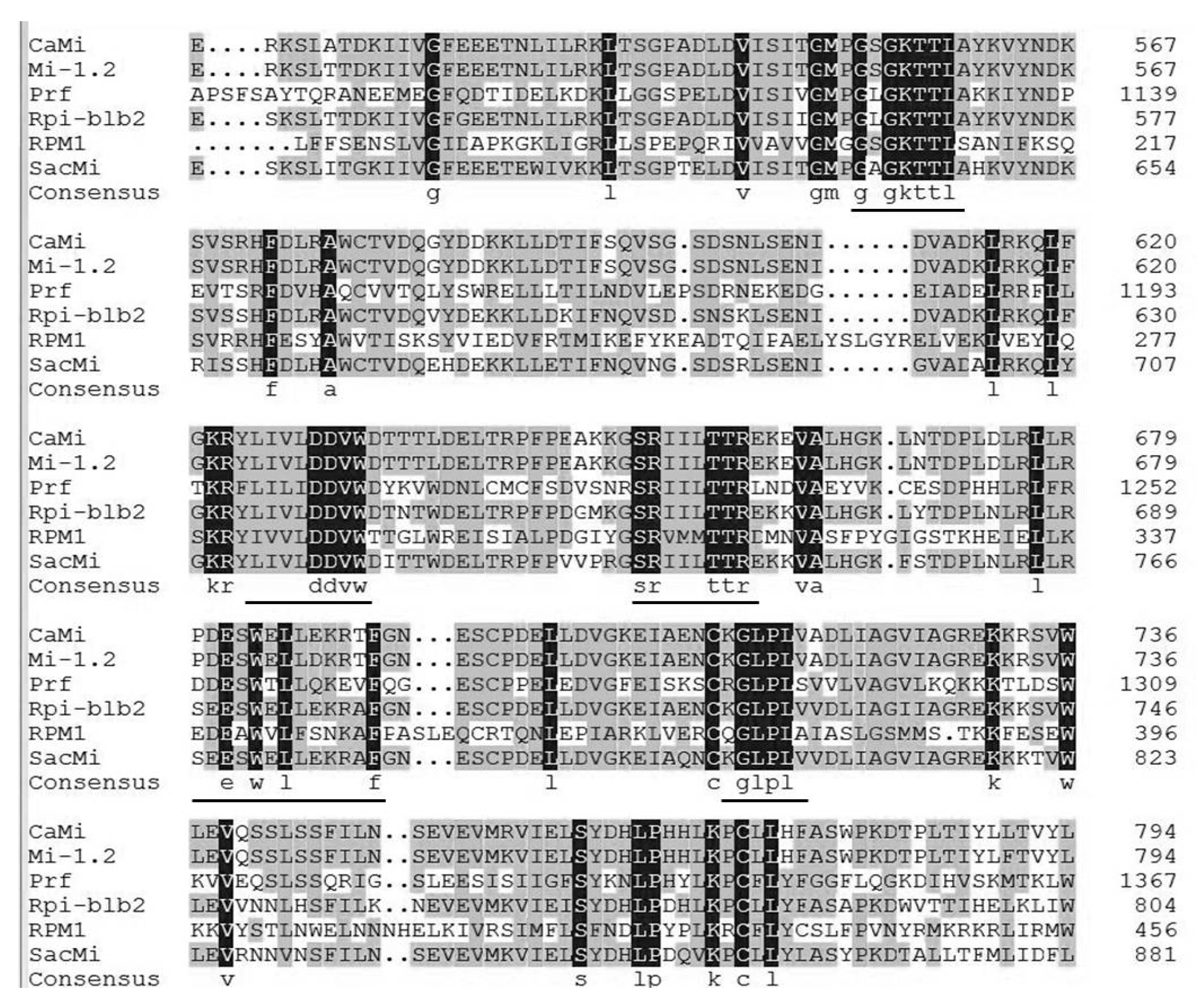
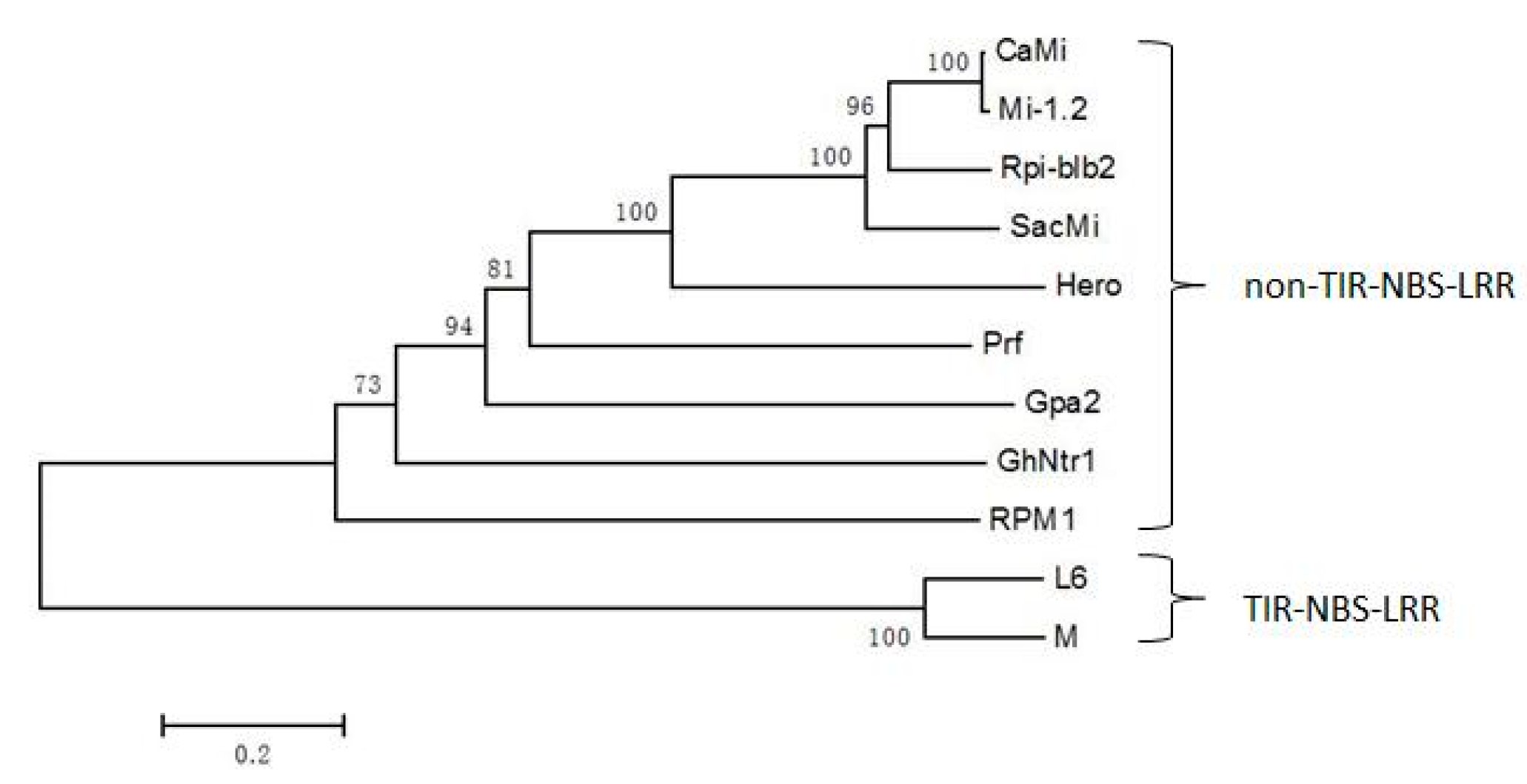
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
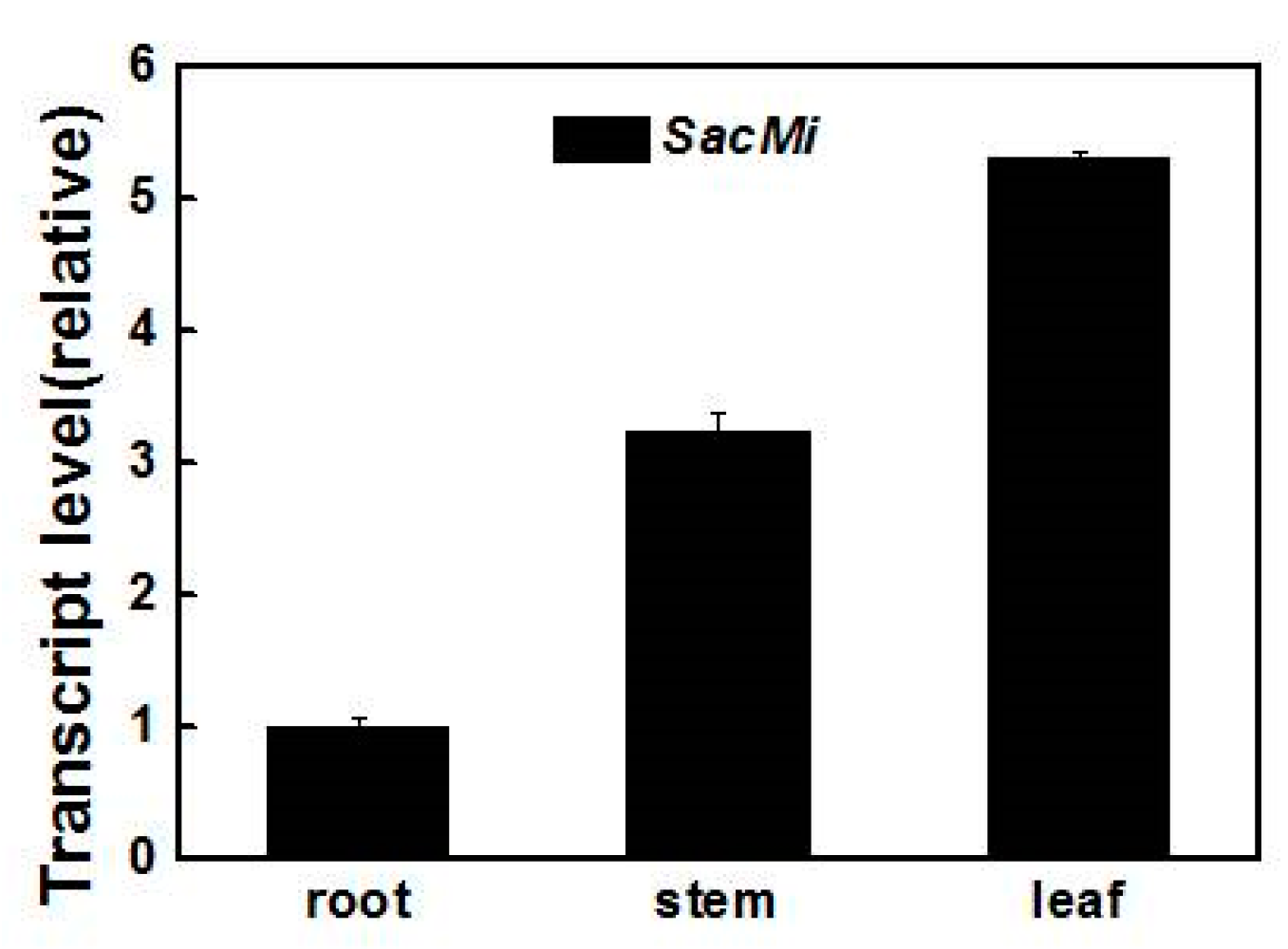
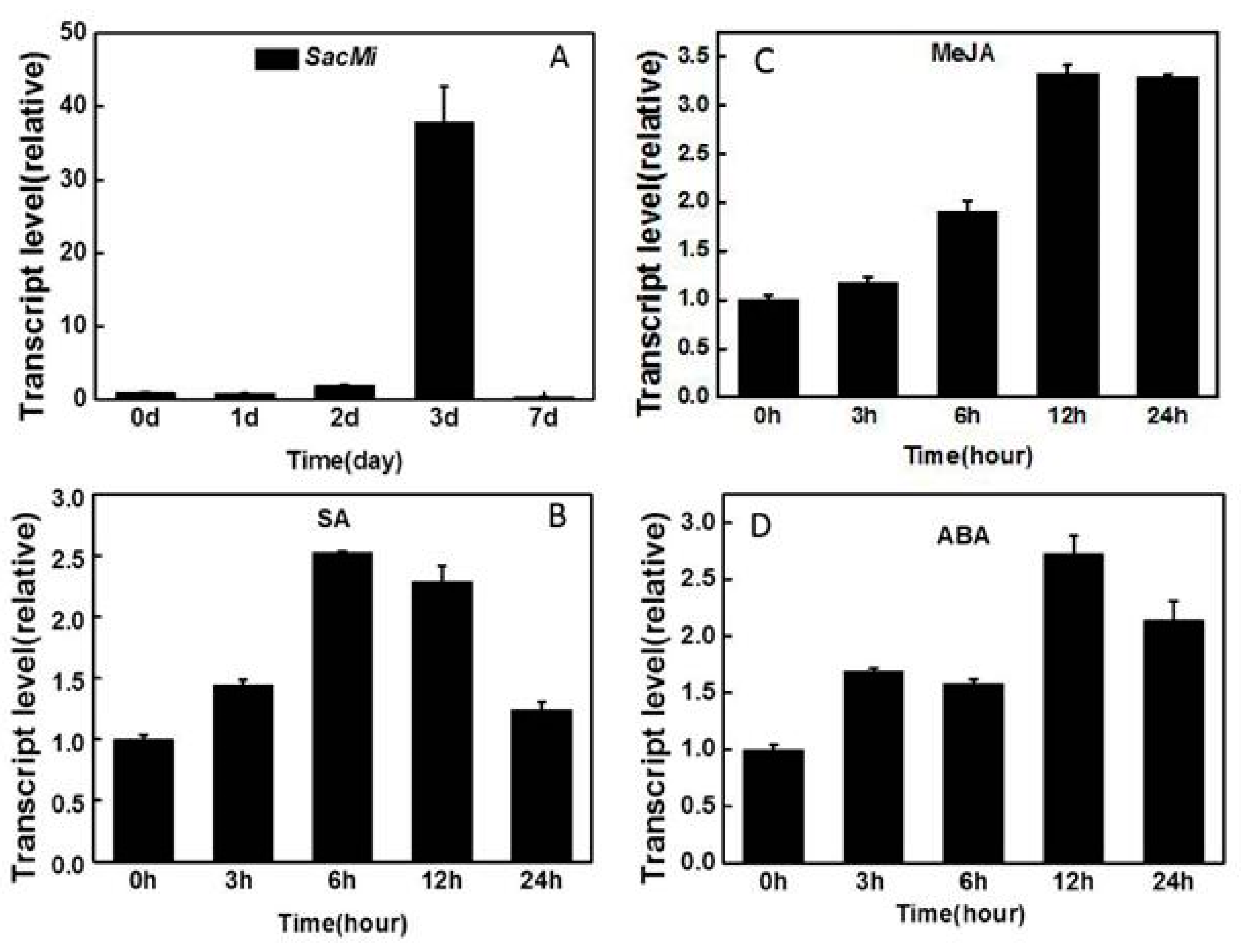
2.3. Expression Analysis of SacMi
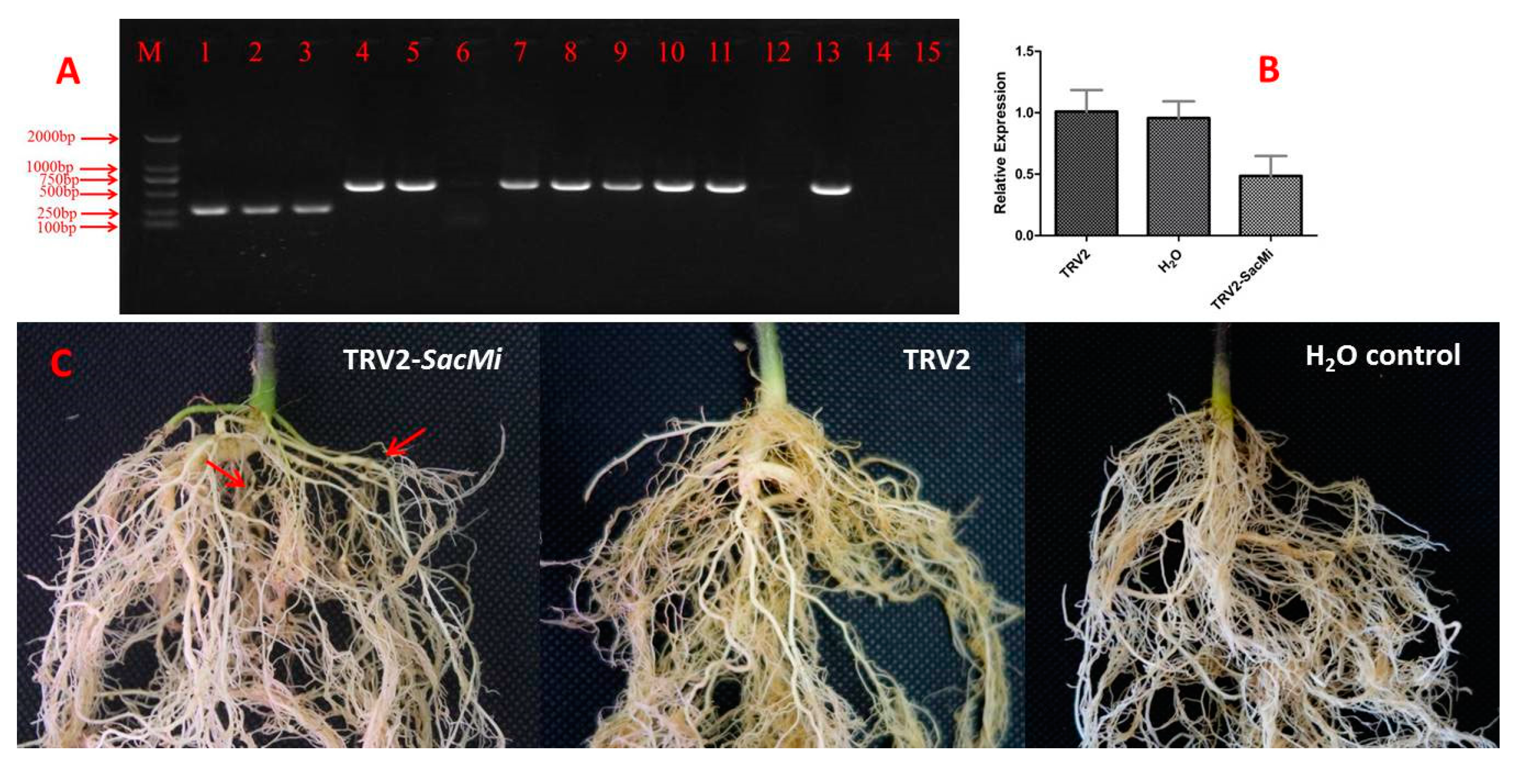
2.4. Silencing of SacMi in S. aculeatissmum Using TRV-VIGS
3. Discussion
4. Materials and Methods
4.1. Full-Length cDNA Cloning
4.2. Bioinformatics Analysis
4.3. Plant Material, Pathogen Inoculation, and Hormone Treatments
4.4. Quantitative Real-Time PCR Analysis
4.5. VIGS of SacMi in S. aculeatissimum
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| NBS | Nucleotide binding site |
| LRR | Leucine-rich repeat |
| TIR | Toll/Interleukin-1 receptor |
| TRV | Tobacco rattle virus |
| VIGS | Virus-induced gene silencing |
| ORF | Open reading frame |
| qRT-PCR | Quantitative real-time PCR |
| SA | Salicylic acid |
| MeJA | Methyl jasmonate |
| ABA GA | Abscisic acid Gibberellic acid |
| RACE | Rapid-amplification of cDNA ends |
References
- Castagnone-Sereno, P. Genetic variability and adaptive evolution in parthenogenetic root-knot nematodes. Heredity 2006, 96, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Djian-Caporalino, C.; Fazari, A.; Arguel, M.J.; Vernie, T.; VandeCasteele, C.; Faure, I.; Brunoud, G.; Pijarowski, L.; Palloix, A.; Lefebvre, V.; et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor. Appl. Genet. 2007, 114, 473–486. [Google Scholar] [CrossRef] [PubMed]
- Walters, S.A.; Barker, K.R. Current distribution of five major Meloidogyne species in the United States. Plant Dis. 1994, 78, 772–774. [Google Scholar] [CrossRef]
- Trudgill, D.L. Parthenogenetic root-knot nematodes (Meloidogyne spp.); how can these biotrophic endoparasites have such an enormous host range? Plant Pathol. 1997, 46, 26–32. [Google Scholar] [CrossRef]
- Milligan, S.B.; Bodeau, J.; Yaghoobi, J.; Kaloshian, I.; Zabel, P.; Williamson, V.M. The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 1998, 10, 1307–1319. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.G.; Li, H.X.; Zhang, L.Y.; Zhang, J.H.; Xiao, J.H.; Ye, Z.B. CaMi, a root-knot nematode resistance gene from hot pepper (Capsium annum L.) confers nematode resistance in tomato. Plant Cell Rep. 2007, 26, 895–905. [Google Scholar] [CrossRef] [PubMed]
- Williamson, V.M.; Hussey, R.S. Nematode pathogenesis and resistance in plants. Plant Cell 1996, 8, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Williamson, V.M.; Kumar, A. Nematode resistance in plants: The battle underground. Trends Genet. 2006, 22, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Hinchliffe, D.; Cantrell, R.; Wang, C.Y.; Roberts, P.; Zhang, J. Identification of molecular markers associated with root-knot nematode resistance in upland cotton. Crop Sci. 2007, 47, 951–960. [Google Scholar] [CrossRef]
- Williamson, V.M. Root-knot nematode resistance genes in tomato and their potential for future use. Annu. Rev. Phytopathol. 1998, 36, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Nombela, G.; Williamson, V.M.; Muniz, M. The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol. Plant Microbe Interact. 2003, 16, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Collonnier, C.; Fock, I.; Kashyap, V.; Rotino, G.L.; Daunay, M.C.; Lian, Y.; Mariska, I.K.; Rajam, M.V.; Servaes, A.; Ducreux, G.; et al. Applications of biotechnology in eggplant. Plant Cell Tissue Organ Cult. 2001, 65, 91–107. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhou, X.H.; Wang, S.B. Genetic diversity of NBS-LRR class disease-resistance gene analogs in cultivated and wild eggplants. Plant Syst. Evol. 2012, 298, 1399–1406. [Google Scholar] [CrossRef]
- Jain, R.K.; Mathur, K.N.; Singh, R.V. Estimation of losses due to plant parasitic nematodes on different crops in India. Indian J. Nematol. 2007, 37, 219–221. [Google Scholar]
- Kinloch, R.A.; Hiebsch, C.K.; Peacock, A.A. Comparative root-knot galling and yield responses of soybean cultivars to Meloidogyne incognita. Plant Dis. 1985, 69, 334–336. [Google Scholar]
- Jacquet, M.; Bongiovanni, M.; Martinez, M.; Verschave, P.; Wajnberg, E.; Castagnone-Sereno, P. Variation in resistance to the root-knot nematode Meloidogyne incognita in tomato genotypes bearing the Mi gene. Plant Pathol. 2005, 54, 93–99. [Google Scholar] [CrossRef]
- Bagnaresi, P.; Sala, T.; Irdani, T.; Scotto, C.; Lamontanara, A.; Beretta, M.; Rotino, G.L.; Sestili, S.; Cattivelli, L.; Sabatini, E. Solanum torvum responses to the root-knot nematode Meloidogyne incognita. BMC Genom. 2013, 14, 540. [Google Scholar] [CrossRef] [PubMed]
- Handique, A.K. Breeding behavior of Solanum khasianum Clarke. Euphytica 1986, 35, 631–632. [Google Scholar] [CrossRef]
- Borua, P.K. Failure in an interspecific cross between Solanum khasianum Clarke and Solanum mammosum L. Euphytica 1990, 46, 1–6. [Google Scholar] [CrossRef]
- Zhou, X.H.; Bao, S.Y.; Liu, J.; Zhuang, Y. De novo sequencing and analysis of the transcriptome of the wild eggplant species Solanum aculeatissimum in response to Verticillium dahliae. Plant Mol. Biol. Rep. 2016, 34, 1193–1203. [Google Scholar] [CrossRef]
- Rattan, P.; Kumar, S.; Salgotra, R.K.; Samnotra, R.K.; Sharma, F. Development of interspecific F1 hybrids (Solanum melongena × Solanum Khasianum) in eggplant through embryo rescue technique. Plant Cell Tissue Organ Cult. 2015, 120, 379–386. [Google Scholar] [CrossRef]
- Zhou, X.H.; Bao, S.Y.; Liu, J.; Zhuang, Y. Production and characterization of an amphidiploid derived from interspecific hybridization between Solanum melongena L. and Solanum aculeatissimum Jacq. Sci. Hortic. 2018, 230, 102–106. [Google Scholar] [CrossRef]
- Pathogen Receptor Genes Database. Available online: http://www. prgdb.org (accessed on 8 April 2017).
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-Encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.J.; Zhao, Z.G.; Malik, A.A.; Qian, C.T.; Chen, J.F. Identification and characterization of potential NBS-encoding resistance genes and induction kinetics of a putative candidate gene associated with downy mildew resistance in Cucumis. BMC Plant Biol. 2010, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, X.H.; Feng, C.; Wu, W.; Zhuang, Y. The function confirmation of Ve gene homolog in S. linnaeanum related to Verticillium wilt resistance by VIGS. Acta Agric. Boreali-Sin. 2014, 29, 217–221. [Google Scholar]
- Tanksley, S.D.; McCouch, S. Seed banks and molecular maps: Unlocking the genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Michelmore, R.W. The impact zone: Genomics and breeding for durable disease resistance. Curr. Opin. Plant Biol. 2003, 6, 397–404. [Google Scholar] [CrossRef]
- Van der Biezen, E.; Jones, J. The NB-ARC domain: A novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, 226–227. [Google Scholar] [CrossRef]
- Aravind, L.; Dixit, V.; Koonin, E. The domains of death: Evolution of the apoptosis machinery. Trends Biol. Sci. 1999, 24, 47–53. [Google Scholar] [CrossRef]
- De Young, B.J.; Inners, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Ernst, K.; Amar, K.; Doris, K. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 2002, 31, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Van der Vossen, E.A.G.; van der Voort, J.; Kanyuka, K.; Bendahmane, A.; Sandbrink, H.; Baulcombe, D.C.; Bakker, J.; Stiekema, J.; Klein-Lankhorst, R.M. Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: A virus and a nematode. Plant J. 2000, 23, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Paal, J.; Henselewski, H.; Muth, J.; Meksem, K.; Menendez, C.M.; Salamini, F.; Ballvora, A.; Gebhardt, C. Molecular cloning of the potato Gro1-4 gene conferring resistance to pathotype Ro1 of the root cyst nematode Globodera rostochiensis, based on a candidate gene approach. Plant J. 2004, 38, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Lagudah, E.S.; Moullet, O.; Appels, R. Map based cloning of a gene sequence encoding a nucleotide binding domain and leucine rich region at the Cre3 nematode resistance locus of wheat. Genome 1997, 40, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Thurau, T.; Sirak, K.; Jung, C.; Cai, D. The promoter of the nematode resistance gene Hs1pro−1 activates a nematode-responsive and feeding site-specific gene expression in sugar beet (Beta vulgaris L.) and Arabidopsis thaliana. Plant Mol. Biol. 2003, 52, 643–660. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, S.; Yamanouchi, U.; Katayose, Y.; Toki, S.; Wang, Z.X.; Kono, I.; Kurata, N.; Yano, M.; Iwata, N.; Sasaki, T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. USA 1998, 95, 1663–1669. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Iwamoto, M.; Monna, L.; Hayasaka, H.; Katayose, Y.; Sasaki, T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999, 19, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.X.; Yano, M.; Yamanouchi, U.; Katayose, Y.; Iwamoto, M.; Sasaki, T.; Yano, M. Expression of the Pib rice-blast-resistance gene family is up-regulated by environmental conditions favoring infection and by chemical signals that trigger secondary plant defences. Plant Mol. Biol. 2001, 47, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defense responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, B.N.; Brooks, D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002, 5, 325–331. [Google Scholar] [CrossRef]
- Flors, V.; Ton, J.; van Doorn, R.; Jakab, G.; Garcia-Agustin, P.; Mauch-Mani, B. Interplay between JA, SA and ABA signaling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 2007, 1, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Branch, C.; Hwang, C.F.; Navarre, D.A.; Williamson, V.M. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol. Plant-Microbe Interact. 2004, 17, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, K.K.; Xie, Q.G.; Mantelin, S.; Bishnoi, U.; Girke, T.; Navarre, D.A.; Kaloshian, I. Tomato susceptibility to root-knot nematodes requires an intact jasmonic acid signaling pathway. Mol. Plant-Microbe Interact. 2008, 21, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Shirano, Y.; Kachroo, P.; Shah, J.; Klessig, D.F. A Gain-of-Function mutation in an Arabidopsis Toll Interleukin 1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell 2002, 14, 3149–3162. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.G.; Kleine, M.; Kifle, S. Positional cloning of a gene for nematode resistance in sugar beet. Science 1997, 275, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Ammati, M.; Thomason, I.; McKinney, H. Retention of resistance to Meloidogyne incognita in Lycopersicon genotypes at high soil temperature. J. Nematol. 1986, 18, 491–495. [Google Scholar] [PubMed]
- Hare, W.W. Resistance in pepper to Meoidogyne inncognita. Phytopathology 1956, 46, 98–104. [Google Scholar]
- Griffin, G.D. Effects of temperature on Meloidogyne hapla in alfalfa. Phytopathology 1969, 59, 599–602. [Google Scholar]
- Jatala, P.; Russell, C.C. Nature of sweet potato resistance to Meloidogyne incognita and the effects of temperature on parasitism. J. Nematol. 1972, 4, 1–7. [Google Scholar] [PubMed]
- Carter, W.W. Influence of soil temperature on Meloidogyne incognita resistant and susceptible cotton, Gossypium hirsutum. J. Nematol. 1982, 14, 343–346. [Google Scholar] [PubMed]
- Rossi, M.; Goggin, F.L.; Milligan, S.B.; Kaloshian, I.; Ullman, D.E.; Williamson, V.W. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc. Natl. Acad. Sci. USA 1998, 95, 9750–9754. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov/ (accessed on 7 April 2017).
- Conserved Domain Database. Available online: https://www.ncbi.nlm.nih.gov/structure/cdd/wrpsb.cgi (accessed on 7 April 2017).
- ExPASy. Available online: http://www.expasy.org/tools (accessed on 18 April 2017).
- Lupas, A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996, 266, 513–525. [Google Scholar] [PubMed]
- EMBnet Node Switzerland. Available online: http://www.ch.embnet.org (accessed on 14 August 2017).
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA 6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Molecular Evolutionary Genetics Analysis. Available online: http://www.megasoftware.net (accessed on 18 April 2017).
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Hussey, R.S.; Barker, K.R. A comparison of methods of collecting inocula of Meloidogyne spp. including a new technique. Plant Dis. Rep. 1973, 57, 1025–1028. [Google Scholar]
- Thies, J.A.; Ariss, J.J. Comparison between the N and Me3 genes conferring resistance to the root-knot nematode (Meloidogyne incognita) in genetically different pepper lines (Capsicum annuum). Eur. J. Plant Pathol. 2009, 125, 545–550. [Google Scholar] [CrossRef]
- Rozen, S.; Skaletsky, H. Primer 3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [PubMed]
- Primer3web. Available online: http://primer3.ut.ee (accessed on 18 April 2017).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]

| PCR Types | Primers | Primer Sequences (5′-3′) |
|---|---|---|
| 5′ RACE RT | SacMi-R2 | GATTTCTCTTCTAAGTCGCTAA |
| 5′ RACE 1st round RT-PCR | SacMi-R3 | TGTTTCGAGCCCCTGGAGTGCT |
| 5′ RACE 2nd round RT-PCR | SacMi-R4 | TCAGCATGATACTTGGATAGAT |
| 5′ RACE 2nd round RT-PCR | SacMi-R5 | TGACGCAACCATTCACCATCAACCTA |
| 3′ RACE 1st round RT-PCR | SacMi-F2 | TTTCGATCATTGGTATGCCGGGTG |
| DNA sequence amplification | SacMi-F3ORF | ATGGAAAGAGACAAAAGGGAAGC |
| DNA sequence amplification | SacMi-R6ORF | CTAATTAAATAATGGGATATTCATC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Liu, J.; Bao, S.; Yang, Y.; Zhuang, Y. Molecular Cloning and Characterization of a Wild Eggplant Solanum aculeatissimum NBS-LRR Gene, Involved in Plant Resistance to Meloidogyne incognita. Int. J. Mol. Sci. 2018, 19, 583. https://doi.org/10.3390/ijms19020583
Zhou X, Liu J, Bao S, Yang Y, Zhuang Y. Molecular Cloning and Characterization of a Wild Eggplant Solanum aculeatissimum NBS-LRR Gene, Involved in Plant Resistance to Meloidogyne incognita. International Journal of Molecular Sciences. 2018; 19(2):583. https://doi.org/10.3390/ijms19020583
Chicago/Turabian StyleZhou, Xiaohui, Jun Liu, Shengyou Bao, Yan Yang, and Yong Zhuang. 2018. "Molecular Cloning and Characterization of a Wild Eggplant Solanum aculeatissimum NBS-LRR Gene, Involved in Plant Resistance to Meloidogyne incognita" International Journal of Molecular Sciences 19, no. 2: 583. https://doi.org/10.3390/ijms19020583





