When Immune Cells Turn Bad—Tumor-Associated Microglia/Macrophages in Glioma
Abstract
:1. Introduction
2. Origin and Physiological Function of Microglia and Macrophages of the Central Nervous System
3. Microglia Models for Functional Studies
4. Activation and Polarization of Resting Microglia and Macrophages
5. Microglia/Macrophages—Glioma Cell Crosstalk
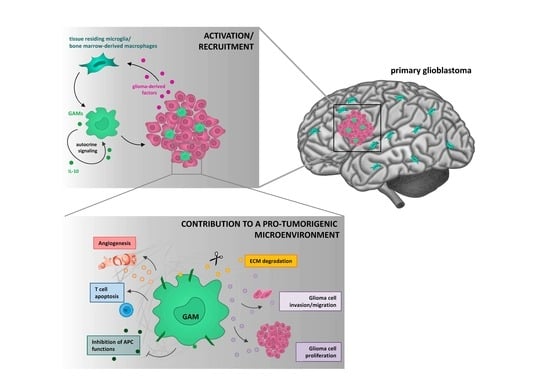
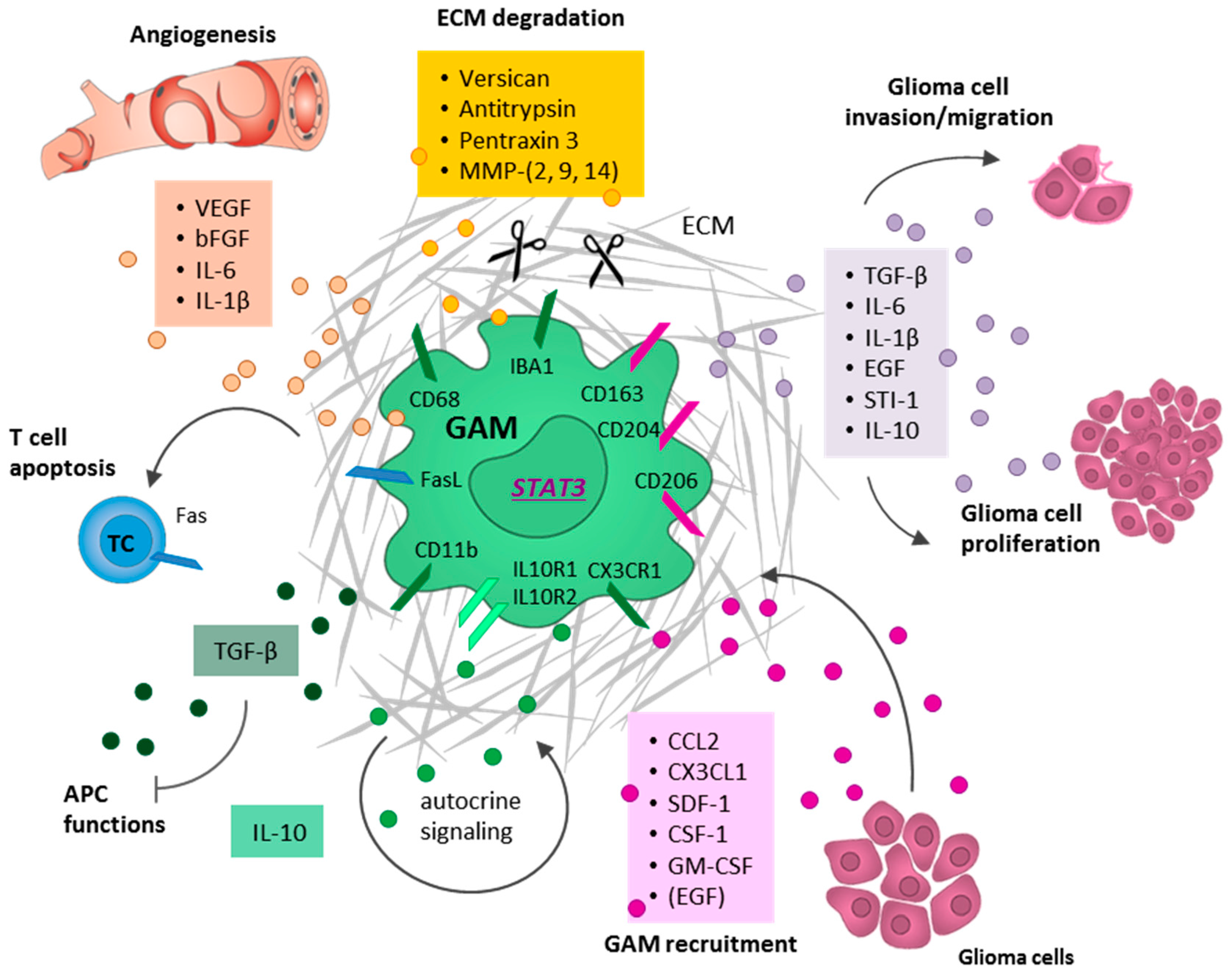
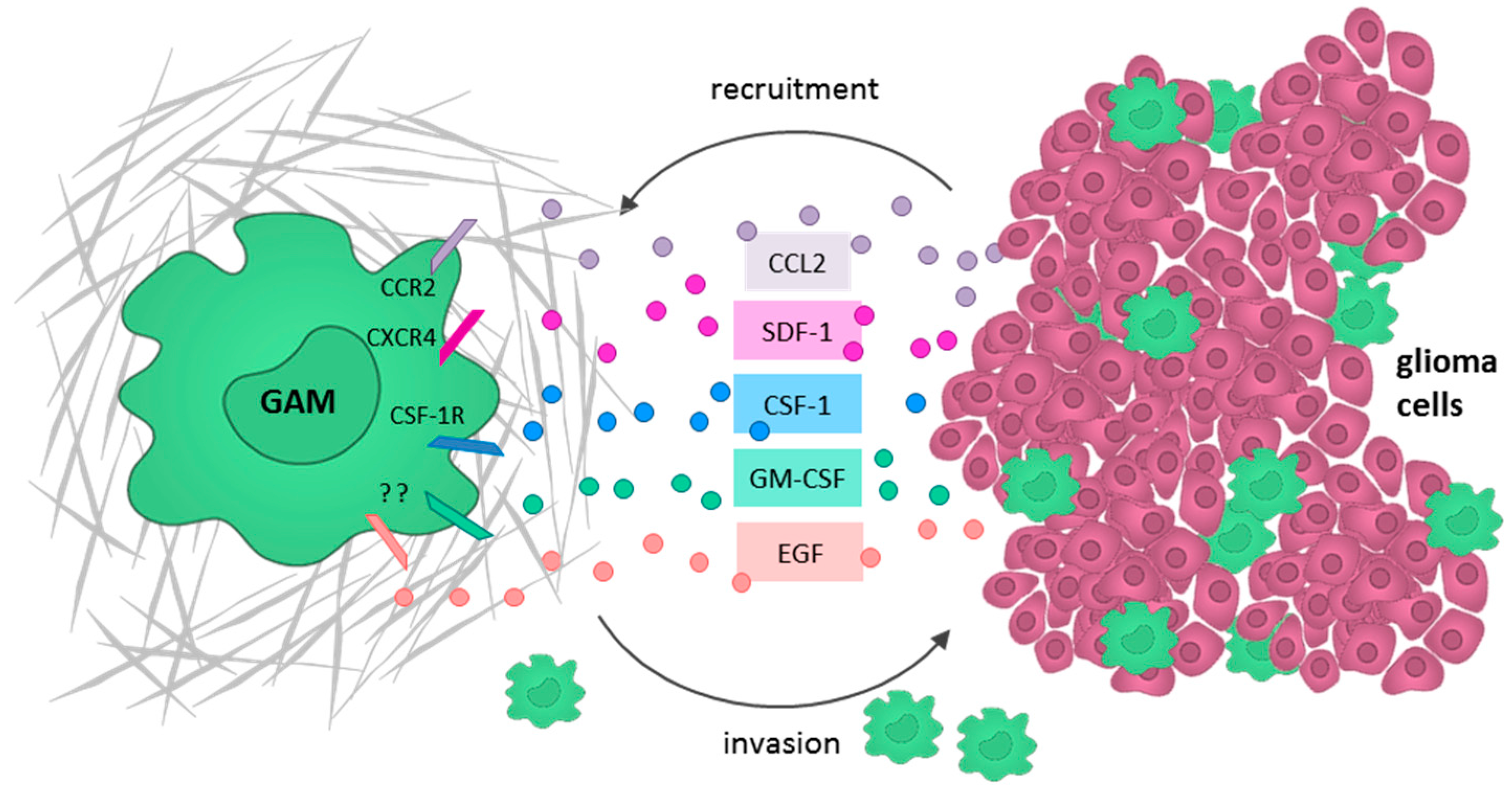
5.1. Glioma Cells Actively Recruit GAMs and Induce a M2-Like Polarization
5.2. GAMs Promote Glioma Cell Invasion and Tumor Growth
5.3. GAMs Affect Neoangiogenesis by Destructing the Extracellular Matrix
5.4. GAMs Contribute to an Immunosuppressive TME
6. Targeting GAMs to Reinforce the Anti-Tumor Immunity
7. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- DeWitt, J.C.; Mock, A.; Louis, D.N. The 2016 WHO classification of central nervous system tumors: What neurologists need to know. Curr. Opin. Neurol. 2017, 30, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. The Microenvironmental Landscape of Brain Tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Haddad-Tóvolli, R.; Dragano, N.R.V.; Ramalho, A.F.S.; Velloso, L.A. Development and Function of the Blood-Brain Barrier in the Context of Metabolic Control. Front. Neurosci. 2017, 11, 224. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Priller, J. The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci. 2017, 20, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Raes, G.; Van den Bergh, R.; De Baetselier, P.; Ghassabeh, G.H.; Scotton, C.; Locati, M.; Mantovani, A.; Sozzani, S. Arginase-1 and Ym1 Are Markers for Murine, but Not Human, Alternatively Activated Myeloid Cells. J. Immunol. 2005, 174, 6561–6562. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Graeber, M.B.; Scheithauer, B.W.; Kreutzberg, G.W. Microglia in brain tumors. Glia 2002, 40, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Poon, C.C.; Sarkar, S.; Yong, V.W.; Kelly, J.J.P. Glioblastoma-associated microglia and macrophages: Targets for therapies to improve prognosis. Brain J. Neurol. 2017, 140, 1548–1560. [Google Scholar] [CrossRef] [PubMed]
- Davies, L.C.; Jenkins, S.J.; Allen, J.E.; Taylor, P.R. Tissue-resident macrophages. Nat. Immunol. 2013, 14, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Greter, M.; Leboeuf, M.; Nandi, S.; See, P.; Gokhan, S.; Mehler, M.F.; Conway, S.J.; Ng, L.G.; Stanley, E.R.; et al. Fate Mapping Analysis Reveals That Adult Microglia Derive from Primitive Macrophages. Science 2010, 330, 841–845. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C.; et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013, 16, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Casano, A.M.; Peri, F. Microglia: Multitasking specialists of the brain. Dev. Cell 2015, 32, 469–477. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Shen, Q.; Xu, P.; Luo, J.J.; Tang, Y. Phagocytosis of microglia in the central nervous system diseases. Mol. Neurobiol. 2014, 49, 1422–1434. [Google Scholar] [CrossRef] [PubMed]
- Gomez Perdiguero, E.; Klapproth, K.; Schulz, C.; Busch, K.; Azzoni, E.; Crozet, L.; Garner, H.; Trouillet, C.; de Bruijn, M.F.; Geissmann, F.; et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 2015, 518, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Hickey, W.; Kimura, H. Perivascular microglial cells of the CNS are bone marrow-derived and present antigen in vivo. Science 1988, 239, 290–292. [Google Scholar] [CrossRef] [PubMed]
- Epelman, S.; Lavine, K.J.; Randolph, G.J. Origin and functions of tissue macrophages. Immunity 2014, 41, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, K.D.; Dickson, D.W.; Rashbaum, W.K.; Lyman, W.D. Localization of morphologically distinct microglial populations in the developing human fetal brain: Implications for ontogeny. Brain Res. Dev. Brain Res. 1990, 55, 95–102. [Google Scholar] [CrossRef]
- Prinz, M.; Priller, J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014, 15, 300–312. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.L.; Bennett, F.C.; Liddelow, S.A.; Ajami, B.; Zamanian, J.L.; Fernhoff, N.B.; Mulinyawe, S.B.; Bohlen, C.J.; Adil, A.; Tucker, A.; et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 2016, 113, E1738–E1746. [Google Scholar] [CrossRef] [PubMed]
- Sousa, C.; Biber, K.; Michelucci, A. Cellular and Molecular Characterization of Microglia: A Unique Immune Cell Population. Front. Immunol. 2017, 8, 198. [Google Scholar] [CrossRef] [PubMed]
- Korzhevskii, D.E.; Kirik, O.V. Brain Microglia and Microglial Markers. Neurosci. Behav. Physiol. 2016, 46, 284–290. [Google Scholar] [CrossRef]
- Badie, B.; Schartner, J.M. Flow Cytometric Characterization of Tumor-associated Macrophages in Experimental Gliomas. Neurosurgery 2000, 46, 957–962. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Brandenburg, S.; Turkowski, K.; Müller, S.; Vajkoczy, P. Resident microglia, and not peripheral macrophages, are the main source of brain tumor mononuclear cells. Int. J. Cancer 2014, 137, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Satoh, J.; Kino, Y.; Asahina, N.; Takitani, M.; Miyoshi, J.; Ishida, T.; Saito, Y. TMEM119 marks a subset of microglia in the human brain. Neuropathology 2016, 36, 39–49. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.D.; de Vellis, J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980, 85, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Giulian, D.; Baker, T.J. Characterization of ameboid microglia isolated from developing mammalian brain. J. Neurosci. 1986, 6, 2163–2178. [Google Scholar] [PubMed]
- Kees, T.; Lohr, J.; Noack, J.; Mora, R.; Gdynia, G.; Tödt, G.; Ernst, A.; Radlwimmer, B.; Falk, C.S.; Herold-Mende, C.; et al. Microglia isolated from patients with glioma gain antitumor activities on poly (I:C) stimulation. Neuro-Oncology 2011, 14, 64–78. [Google Scholar] [CrossRef] [PubMed]
- Blasi, E.; Barluzzi, R.; Bocchini, V.; Mazzolla, R.; Bistoni, F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J. Neuroimmunol. 1990, 27, 229–237. [Google Scholar] [CrossRef]
- Janabi, N.; Peudenier, S.; Héron, B.; Ng, K.H.; Tardieu, M. Establishment of human microglial cell lines after transfection of primary cultures of embryonic microglial cells with the SV40 large T antigen. Neurosci. Lett. 1995, 195, 105–108. [Google Scholar] [CrossRef]
- Nagai, A.; Nakagawa, E.; Hatori, K.; Choi, H.B.; McLarnon, J.G.; Lee, M.A.; Kim, S.U. Generation and characterization of immortalized human microglial cell lines: Expression of cytokines and chemokines. Neurobiol. Dis. 2001, 8, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Henn, A.; Lund, S.; Hedtjärn, M.; Schrattenholz, A.; Pörzgen, P.; Leist, M. The suitability of BV2 cells as alternative model system for primary microglia cultures or for animal experiments examining brain inflammation. ALTEX 2009, 26, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Kim, S.H.; Arifuzzaman, S.; Yoon, T.; Chai, J.C.; Lee, Y.S.; Park, K.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing reveals that LPS-triggered transcriptional responses in established microglia BV2 cell lines are poorly representative of primary microglia. J. Neuroinflamm. 2016, 13, 182. [Google Scholar] [CrossRef] [PubMed]
- Horvath, R.J.; Nutile-McMenemy, N.; Alkaitis, M.S.; Deleo, J.A. Differential migration, LPS-induced cytokine, chemokine, and NO expression in immortalized BV-2 and HAPI cell lines and primary microglial cultures. J. Neurochem. 2008, 107, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Marek, R.; Caruso, M.; Rostami, A.; Grinspan, J.B.; Das Sarma, J. Magnetic cell sorting: A fast and effective method of concurrent isolation of high purity viable astrocytes and microglia from neonatal mouse brain tissue. J. Neurosci. Methods 2008, 175, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Losciuto, S.; Dorban, G.; Gabel, S.; Gustin, A.; Hoenen, C.; Grandbarbe, L.; Heuschling, P.; Heurtaux, T. An efficient method to limit microglia-dependent effects in astroglial cultures. J. Neurosci. Methods 2012, 207, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, T.; Park, K.C.; Toyonaga, S.; Yamada, S.M.; Nakabayashi, H.; Nakai, E.; Ikawa, N.; Furuya, M.; Tominaga, A.; Shimizu, K. Characterization of microglia induced from mouse embryonic stem cells and their migration into the brain parenchyma. J. Neuroimmunol. 2005, 160, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, G. Embryoid body formation from human pluripotent stem cells in chemically defined E8 media. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2008. [Google Scholar]
- Jung, S.; Aliberti, J.; Graemmel, P.; Sunshine, M.J.; Kreutzberg, G.W.; Sher, A.; Littman, D.R. Analysis of Fractalkine Receptor CX3CR1 Function by Targeted Deletion and Green Fluorescent Protein Reporter Gene Insertion. Mol. Cell. Biol. 2000, 20, 4106–4114. [Google Scholar] [CrossRef] [PubMed]
- London, A.; Benhar, I.; Mattapallil, M.J.; Mack, M.; Caspi, R.R.; Schwartz, M. Functional macrophage heterogeneity in a mouse model of autoimmune central nervous system pathology. J. Immunol. Baltim. 2013, 190, 3570–3578. [Google Scholar] [CrossRef] [PubMed]
- Ransohoff, R.M.; Perry, V.H. Microglial physiology: Unique stimuli, specialized responses. Annu. Rev. Immunol. 2009, 27, 119–145. [Google Scholar] [CrossRef] [PubMed]
- Humpel, C. Organotypic brain slice cultures: A review. Neuroscience 2015, 305, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Van Rooijen, N.; Hendrikx, E. Liposomes for specific depletion of macrophages from organs and tissues. Methods Mol. Biol. 2010, 605, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Nimmerjahn, A.; Kirchhoff, F.; Helmchen, F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 2005, 308, 1314–1318. [Google Scholar] [CrossRef] [PubMed]
- Bayerl, S.H.; Niesner, R.; Cseresnyes, Z.; Radbruch, H.; Pohlan, J.; Brandenburg, S.; Czabanka, M.A.; Vajkoczy, P. Time lapse in vivo microscopy reveals distinct dynamics of microglia-tumor environment interactions—A new role for the tumor perivascular space as highway for trafficking microglia. Glia 2016, 64, 1210–1226. [Google Scholar] [CrossRef] [PubMed]
- Dailey, M.E.; Waite, M. Confocal imaging of microglial cell dynamics in hippocampal slice cultures. Methods 1999, 18, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Kurpius, D.; Nolley, E.P.; Dailey, M.E. Purines induce directed migration and rapid homing of microglia to injured pyramidal neurons in developing hippocampus. Glia 2007, 55, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Boehm, U.; Klamp, T.; Groot, M.; Howard, J.C. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997, 15, 749–795. [Google Scholar] [CrossRef] [PubMed]
- Colton, C.A.; Wilcock, D.M. Assessing activation states in microglia. CNS Neurol. Disord. Drug Targets 2010, 9, 174–191. [Google Scholar] [CrossRef] [PubMed]
- Varnum, M.M.; Ikezu, T. The classification of microglial activation phenotypes on neurodegeneration and regeneration in Alzheimer’s disease brain. Arch. Immunol. Ther. Exp. (Warsz.) 2012, 60, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Boche, D.; Perry, V.H.; Nicoll, J.A. Review: Activation patterns of microglia and their identification in the human brain. Neuropathol. Appl. Neurobiol. 2013, 39, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Orihuela, R.; McPherson, C.A.; Harry, G.J. Microglial M1/M2 polarization and metabolic states. Br. J. Pharmacol. 2016, 173, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef]
- Shaked, I.; Porat, Z.; Gersner, R.; Kipnis, J.; Schwartz, M. Early activation of microglia as antigen-presenting cells correlates with T cell-mediated protection and repair of the injured central nervous system. J. Neuroimmunol. 2004, 146, 84–93. [Google Scholar] [CrossRef] [PubMed]
- MacMicking, J.; Xie, Q.W.; Nathan, C. Nitric oxide and macrophage function. Annu. Rev. Immunol. 1997, 15, 323–350. [Google Scholar] [CrossRef] [PubMed]
- Villalta, S.A.; Nguyen, H.X.; Deng, B.; Gotoh, T.; Tidball, J.G. Shifts in macrophage phenotypes and macrophage competition for arginine metabolism affect the severity of muscle pathology in muscular dystrophy. Hum. Mol. Genet. 2009, 18, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Davalos, D.; Grutzendler, J.; Yang, G.; Kim, J.V.; Zuo, Y.; Jung, S.; Littman, D.R.; Dustin, M.L.; Gan, W.-B. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 2005, 8, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Town, T.; Nikolic, V.; Tan, J. The microglial “activation” continuum: From innate to adaptive responses. J. Neuroinflamm. 2005, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C.F.; Murray, H.W.; Wiebe, M.E.; Rubin, B.Y. Identification of interferon-gamma as the lymphokine that activates human macrophage oxidative metabolism and antimicrobial activity. J. Exp. Med. 1983, 158, 670–689. [Google Scholar] [CrossRef] [PubMed]
- Henkel, J.S.; Beers, D.R.; Zhao, W.; Appel, S.H. Microglia in ALS: The good, the bad, and the resting. J. Neuroimmune Pharmacol. 2009, 4, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Colton, C.A. Heterogeneity of microglial activation in the innate immune response in the brain. J. Neuroimmune Pharmacol. 2009, 4, 399–418. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.S.; Masliah, E.; Fox, H.S. CD163 identifies a unique population of ramified microglia in HIV encephalitis (HIVE). J. Neuropathol. Exp. Neurol. 2004, 63, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; VECCHI, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.; Keshav, S.; Harris, N.; Gordon, S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: A marker of alternative immunologic macrophage activation. J. Exp. Med. 1992, 176, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Loke, P.; Nair, M.G.; Parkinson, J.; Guiliano, D.; Blaxter, M.; Allen, J.E. IL-4 dependent alternatively-activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 2002, 3, 7. [Google Scholar] [CrossRef]
- Watanabe, K.; Jose, P.J.; Rankin, S.M. Eotaxin-2 generation is differentially regulated by lipopolysaccharide and IL-4 in monocytes and macrophages. J. Immunol. 2002, 168, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Sawada, M.; Suzumura, A.; Hosoya, H.; Marunouchi, T.; Nagatsu, T. Interleukin-10 inhibits both production of cytokines and expression of cytokine receptors in microglia. J. Neurochem. 1999, 72, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Bonneau, R.H. Stress-induced elevation of glucocorticoids increases microglia proliferation through NMDA receptor activation. J. Neuroimmunol. 2006, 171, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Ummenthum, K.; Peferoen, L.A.N.; Finardi, A.; Baker, D.; Pryce, G.; Mantovani, A.; Bsibsi, M.; Bottazzi, B.; Peferoen-Baert, R.; van der Valk, P.; et al. Pentraxin-3 is upregulated in the central nervous system during MS and EAE, but does not modulate experimental neurological disease. Eur. J. Immunol. 2016, 46, 701–711. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; a Dzaye, O.D.; Hahn, A.; Yu, Y.; Scavetta, R.J.; Dittmar, G.; Kaczmarek, A.K.; Dunning, K.R.; Ricciardelli, C.; Rinnenthal, J.L.; et al. Glioma-derived versican promotes tumor expansion via glioma-associated microglial/macrophages Toll-like receptor 2 signaling. Neuro-Oncology 2014, 17, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Ohnishi, K.; Kuratsu, J.; Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008, 216, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Shemer, A.; Erny, D.; Jung, S.; Prinz, M. Microglia Plasticity during Health and Disease: An Immunological Perspective. Trends Immunol. 2015, 36, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Rosario, A.M.; Cruz, P.E.; Ceballos-Diaz, C.; Strickland, M.R.; Siemienski, Z.; Pardo, M.; Schob, K.-L.; Li, A.; Aslanidi, G.V.; Srivastava, A.; et al. Microglia-specific targeting by novel capsid-modified AAV6 vectors. Mol. Ther. Methods Clin. Dev. 2016, 3, 16026. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Ibata, I.; Ito, D.; Ohsawa, K.; Kohsaka, S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem. Biophys. Res. Commun. 1996, 224, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Gautier, E.L.; Shay, T.; Miller, J.; Greter, M.; Jakubzick, C.; Ivanov, S.; Helft, J.; Chow, A.; Elpek, K.G.; Gordonov, S.; et al. Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012, 13, 1118–1128. [Google Scholar] [CrossRef] [PubMed]
- Prinz, M.; Priller, J.; Sisodia, S.S.; Ransohoff, R.M. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 2011, 14, 1227–1235. [Google Scholar] [CrossRef] [PubMed]
- Gordon, S.; Hamann, J.; Lin, H.-H.; Stacey, M. F4/80 and the related adhesion-GPCRs. Eur. J. Immunol. 2011, 41, 2472–2476. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, M.; Pino, P.A.; Saederup, N.; Charo, I.F.; Ransohoff, R.M.; Cardona, A.E. The Fractalkine Receptor but Not CCR2 Is Present on Microglia from Embryonic Development throughout Adulthood. J. Immunol. 2011, 188, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Szulzewsky, F.; Yerevanian, A.; Chen, Z.; Heinzmann, D.; Rasmussen, R.D.; Alvarez-Garcia, V.; Kim, Y.; Wang, B.; Tamagno, I.; et al. Loss of CX3CR1 increases accumulation of inflammatory monocytes and promotes gliomagenesis. Oncotarget 2015, 6, 15077–15094. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.P.; Zhang, X.; Frauwirth, K.A.; Mosser, D.M. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 2006, 80, 1298–1307. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.C.; Showers, C.R.; Anderson, D.E.; Anderson, L.; Canoll, P.; Bruce, J.N.; Anderson, R.C.E. Tumor-Associated Macrophages in Glioma: Friend or Foe? J. Oncol. 2013, 2013, 486912. [Google Scholar] [CrossRef] [PubMed]
- Verdijk, P.; van Veelen, P.A.; de Ru, A.H.; Hensbergen, P.J.; Mizuno, K.; Koerten, H.K.; Koning, F.; Tensen, C.P.; Mommaas, A.M. Morphological changes during dendritic cell maturation correlate with cofilin activation and translocation to the cell membrane. Eur. J. Immunol. 2004, 34, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Zeiner, P.S.; Preusse, C.; Blank, A.-E.; Zachskorn, C.; Baumgarten, P.; Caspary, L.; Braczynski, A.K.; Weissenberger, J.; Bratzke, H.; Reiß, S.; et al. MIF Receptor CD74 is Restricted to Microglia/Macrophages, Associated with a M1-Polarized Immune Milieu and Prolonged Patient Survival in Gliomas. Brain Pathol. 2014, 25, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Abou-Ghazal, M.; Yang, D.S.; Qiao, W.; Reina-Ortiz, C.; Wei, J.; Kong, L.-Y.; Fuller, G.N.; Hiraoka, N.; Priebe, W.; Sawaya, R.; et al. The Incidence, Correlation with Tumor Infiltrating Inflammation, and Prognosis of p-STAT3 Expression in Human Gliomas. Clin. Cancer Res. 2008, 14, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sarkar, S.; Cua, R.; Zhou, Y.; Hader, W.; Yong, V.W. A dialog between glioma and microglia that promotes tumor invasiveness through the CCL2/CCR2/interleukin-6 axis. Carcinogenesis 2012, 33, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Galea, I.; Palin, K.; Newman, T.A.; Van Rooijen, N.; Perry, V.H.; Boche, D. Mannose receptor expression specifically reveals perivascular macrophages in normal, injured, and diseased mouse brain. Glia 2005, 49, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Raes, G.; De Baetselier, P.; Noël, W.; Beschin, A.; Brombacher, F.; Hassanzadeh Gh, G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. J. Leukoc. Biol. 2002, 71, 597–602. [Google Scholar] [PubMed]
- Desbaillets, I.; Tada, M.; De Tribolet, N.; Diserens, A.-C.; Hamou, M.-F.; Van Meir, E.G. Human astrocytomas and glioblastomas express monocyte chemoattractant protein-1 (MCP-1) in vivo and in vitro. Int. J. Cancer 1994, 58, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Platten, M.; Kretz, A.; Naumann, U.; Aulwurm, S.; Egashira, K.; Isenmann, S.; Weller, M. Monocyte chemoattractant protein-1 increases microglial infiltration and aggressiveness of gliomas. Ann. Neurol. 2003, 54, 388–392. [Google Scholar] [CrossRef] [PubMed]
- Kuratsu, J.; Yoshizato, K.; Yoshimura, T.; Leonard, E.J.; Takeshima, H.; Ushio, Y. Quantitative study of monocyte chemoattractant protein-1 (MCP-1) in cerebrospinal fluid and cyst fluid from patients with malignant glioma. J. Natl. Cancer Inst. 1993, 85, 1836–1839. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Li, W.-J.; Wei, F.-Q.; Wong, T.-S.; Lei, W.-B.; Zhu, X.-L.; Li, J.; Wen, W.-P. Blockade of MCP-1/CCR4 signaling-induced recruitment of activated regulatory cells evokes an antitumor immune response in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 37714–37727. [Google Scholar] [CrossRef] [PubMed]
- Okada, M. Tumor-associated macrophage/microglia infiltration in human gliomas is correlated with MCP-3, but not MCP-1. Int. J. Oncol. 2009, 34, 1621–1627. [Google Scholar] [PubMed]
- Wang, S.-C.; Hong, J.-H.; Hsueh, C.; Chiang, C.-S. Tumor-secreted SDF-1 promotes glioma invasiveness and TAM tropism toward hypoxia in a murine astrocytoma model. Lab. Investig. 2011, 92, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Pyonteck, S.M.; Akkari, L.; Schuhmacher, A.J.; Bowman, R.L.; Sevenich, L.; Quail, D.F.; Olson, O.C.; Quick, M.L.; Huse, J.T.; Teijeiro, V.; et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat. Med. 2013, 19, 1264–1272. [Google Scholar] [CrossRef] [PubMed]
- Sielska, M.; Przanowski, P.; Wylot, B.; Gabrusiewicz, K.; Maleszewska, M.; Kijewska, M.; Zawadzka, M.; Kucharska, J.; Vinnakota, K.; Kettenmann, H.; et al. Distinct roles of CSF family cytokines in macrophage infiltration and activation in glioma progression and injury response. J. Pathol. 2013, 230, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Nolte, C.; Kirchhoff, F.; Kettenmann, H. Epidermal growth factor is a motility factor for microglial cells in vitro: Evidence for EGF receptor expression. Eur. J. Neurosci. 1997, 9, 1690–1698. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Tian, D.; Guo, Z.; Fang, J.; Zhang, Q.; Yu, Z.; Xie, M.; Zhang, H.; Lü, J.; Wang, W. Inhibition of EGFR/MAPK signaling reduces microglial inflammatory response and the associated secondary damage in rats after spinal cord injury. J. Neuroinflamm. 2012, 9, 178. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.-S.; Liu, J.-L.; Li, C.-Y.; Li, X.; Xie, M.-J.; Wang, W.; Tian, D.-S. Rapidly activated epidermal growth factor receptor mediates lipopolysaccharide-triggered migration of microglia. Neurochem. Int. 2015, 90, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Bettinger, I.; Thanos, S.; Paulus, W. Microglia promote glioma migration. Acta Neuropathol. 2002, 103, 351–355. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, S.; Strommer, K.; Frei, K.; Siepl, C.; de Tribolet, N.; Heid, I.; Fontana, A. Immunosuppression and transforming growth factor-beta in glioblastoma. Preferential production of transforming growth factor-beta 2. J. Immunol. 1989, 143, 3222–3229. [Google Scholar] [PubMed]
- Samuels, V.; Barrett, J.M.; Bockman, S.; Pantazis, C.G.; Allen, M.B. Immunocytochemical study of transforming growth factor expression in benign and malignant gliomas. Am. J. Pathol. 1989, 134, 894–902. [Google Scholar] [PubMed]
- Kaminska, B.; Kocyk, M.; Kijewska, M. TGF beta signaling and its role in glioma pathogenesis. Adv. Exp. Med. Biol. 2013, 986, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Wick, W.; Platten, M.; Weller, M. Glioma cell invasion: Regulation of metalloproteinase activity by TGF-beta. J. Neurooncol. 2001, 53, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, R.K.; Sørensen, M.D.; Aaberg-Jessen, C.; Hermansen, S.K.; Kristensen, B.W. Expression and prognostic impact of matrix metalloproteinase-2 (MMP-2) in astrocytomas. PLoS ONE 2017, 12, e0172234. [Google Scholar] [CrossRef] [PubMed]
- Markovic, D.S.; Vinnakota, K.; Chirasani, S.; Synowitz, M.; Raguet, H.; Stock, K.; Sliwa, M.; Lehmann, S.; Kalin, R.; van Rooijen, N.; et al. Gliomas induce and exploit microglial MT1-MMP expression for tumor expansion. Proc. Natl. Acad. Sci. USA 2009, 106, 12530–12535. [Google Scholar] [CrossRef] [PubMed]
- Vinnakota, K.; Hu, F.; Ku, M.C.; Georgieva, P.B.; Szulzewsky, F.; Pohlmann, A.; Waiczies, S.; Waiczies, H.; Niendorf, T.; Lehnardt, S.; et al. Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro-Oncology 2013, 15, 1457–1468. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, A.C.C.; Wang, H.; Fan, H.; Chen, X.; Zhang, I.; Zhang, L.; Lima, F.R.S.; Badie, B. Increased expression of stress inducible protein 1 in glioma-associated microglia/macrophages. J. Neuroimmunol. 2014, 274, 71–77. [Google Scholar] [CrossRef] [PubMed]
- De, I.; Steffen, M.D.; Clark, P.A.; Patros, C.J.; Sokn, E.; Bishop, S.M.; Litscher, S.; Maklakova, V.I.; Kuo, J.S.; Rodriguez, F.J.; et al. CSF1 Overexpression Promotes High-Grade Glioma Formation without Impacting the Polarization Status of Glioma-Associated Microglia and Macrophages. Cancer Res. 2016, 76, 2552–2560. [Google Scholar] [CrossRef] [PubMed]
- Coniglio, S.J.; Eugenin, E.; Dobrenis, K.; Stanley, E.R.; West, B.L.; Symons, M.H.; Segall, J.E. Microglial Stimulation of Glioblastoma Invasion Involves Epidermal Growth Factor Receptor (EGFR) and Colony Stimulating Factor 1 Receptor (CSF-1R) Signaling. Mol. Med. 2012, 18, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.W.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.H.; Xu, A.M.; White, F.M. Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2009, 2, re6. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, S.; Müller, A.; Turkowski, K.; Radev, Y.T.; Rot, S.; Schmidt, C.; Bungert, A.D.; Acker, G.; Schorr, A.; Hippe, A.; et al. Resident microglia rather than peripheral macrophages promote vascularization in brain tumors and are source of alternative pro-angiogenic factors. Acta Neuropathol. 2016, 131, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.; Liang, J.; Holmes, L.; Zurita, A.J.; Henry, V.; Heymach, J.V.; de Groot, J.F. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro-Oncology 2012, 14, 1379–1392. [Google Scholar] [CrossRef] [PubMed]
- Piperi, C.; Samaras, V.; Levidou, G.; Kavantzas, N.; Boviatsis, E.; Petraki, K.; Grivas, A.; Barbatis, C.; Varsos, V.; Patsouris, E.; et al. Prognostic significance of IL-8-STAT-3 pathway in astrocytomas: Correlation with IL-6, VEGF and microvessel morphometry. Cytokine 2011, 55, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Kierdorf, K.; Fritz, G. RAGE regulation and signaling in inflammation and beyond. J. Leukoc. Biol. 2013, 94, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Zhang, I.Y.; Liang, J.; Wang, H.; Ouyang, M.; Wu, S.; da Fonseca, A.C.C.; Weng, L.; Yamamoto, Y.; et al. RAGE Expression in Tumor-Associated Macrophages Promotes Angiogenesis in Glioma. Cancer Res. 2014, 74, 7285–7297. [Google Scholar] [CrossRef] [PubMed]
- Paulus, W.; Baur, I.; Huettner, C.; Schmausser, B.; Roggendorf, W.; Schlingensiepen, K.H.; Brysch, W. Effects of transforming growth factor-beta 1 on collagen synthesis, integrin expression, adhesion and invasion of glioma cells. J. Neuropathol. Exp. Neurol. 1995, 54, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Czub, S.; Greif, M.; Vince, G.H.; Süss, N.; Kerkau, S.; Rieckmann, P.; Roggendorf, W.; Roosen, K.; Tonn, J.C. Microglial/macrophage expression of interleukin 10 in human glioblastomas. Int. J. Cancer 1999, 82, 12–16. [Google Scholar] [CrossRef]
- Kiefer, R.; Supler, M.L.; Toyka, K.V.; Streit, W.J. In situ detection of transforming growth factor-beta mRNA in experimental rat glioma and reactive glial cells. Neurosci. Lett. 1994, 166, 161–164. [Google Scholar] [CrossRef]
- Wesolowska, A.; Kwiatkowska, A.; Slomnicki, L.; Dembinski, M.; Master, A.; Sliwa, M.; Franciszkiewicz, K.; Chouaib, S.; Kaminska, B. Microglia-derived TGF-beta as an important regulator of glioblastoma invasion—An inhibition of TGF-beta-dependent effects by shRNA against human TGF-beta type II receptor. Oncogene 2008, 27, 918–930. [Google Scholar] [CrossRef] [PubMed]
- Suzumura, A.; Sawada, M.; Yamamoto, H.; Marunouchi, T. Transforming growth factor-beta suppresses activation and proliferation of microglia in vitro. J. Immunol. 1993, 151, 2150–2158. [Google Scholar] [PubMed]
- Hussain, S.F.; Yang, D.; Suki, D.; Aldape, K.; Grimm, E.; Heimberger, A.B. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro-Oncology 2006, 8, 261–279. [Google Scholar] [CrossRef] [PubMed]
- Kostianovsky, A.M.; Maier, L.M.; Anderson, R.C.; Bruce, J.N.; Anderson, D.E. Astrocytic Regulation of Human Monocytic/Microglial Activation. J. Immunol. 2008, 181, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Badie, B.; Bartley, B.; Schartner, J. Differential expression of MHC class II and B7 costimulatory molecules by microglia in rodent gliomas. J. Neuroimmunol. 2002, 133, 39–45. [Google Scholar] [CrossRef]
- Frei, K.; Lins, H.; Schwerdel, C.; Fontana, A. Antigen presentation in the central nervous system. The inhibitory effect of IL-10 on MHC class II expression and production of cytokines depends on the inducing signals and the type of cell analyzed. J. Immunol. 1994, 152, 2720–2728. [Google Scholar] [PubMed]
- Zhang, L.; Alizadeh, D.; Van Handel, M.; Kortylewski, M.; Yu, H.; Badie, B. Stat3 inhibition activates tumor macrophages and abrogates glioma growth in mice. Glia 2009, 57, 1458–1467. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Terai, M.; Tamura, Y.; Alexeev, V.; Mastrangelo, M.J.; Selvan, S.R. Interleukin 10 in the tumor microenvironment: A target for anticancer immunotherapy. Immunol. Res. 2011, 51, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Huettner, C.; Czub, S.; Kerkau, S.; Roggendorf, W.; Tonn, J.C. Interleukin 10 is expressed in human gliomas in vivo and increases glioma cell proliferation and motility in vitro. Anticancer Res. 1997, 17, 3217–3224. [Google Scholar] [PubMed]
- Luwor, R.B.; Stylli, S.S.; Kaye, A.H. The role of Stat3 in glioblastoma multiforme. J. Clin. Neurosci. 2013, 20, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Mizoguchi, M.; Betensky, R.A.; Batchelor, T.T.; Bernay, D.C.; Louis, D.N.; Nutt, C.L. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: Correlation with EGFR status, tumor grade, and survival. J. Neuropathol. Exp. Neurol. 2006, 65, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, H.; Zhang, W.; Huang, H.J.; Liao, W.S.L.; Fuller, G.N. Analysis of the activation status of Akt, NFkappaB, and Stat3 in human diffuse gliomas. Lab. Investig. 2004, 84, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.-W.; Cao, X.; Zhu, H.; Ali-Osman, F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin. Cancer Res. 2008, 14, 6042–6054. [Google Scholar] [CrossRef] [PubMed]
- See, A.P.; Han, J.E.; Phallen, J.; Binder, Z.; Gallia, G.; Pan, F.; Jinasena, D.; Jackson, C.; Belcaid, Z.; Jeong, S.J.; et al. The role of STAT3 activation in modulating the immune microenvironment of GBM. J. Neurooncol. 2012, 110, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Iwamaru, A.; Szymanski, S.; Iwado, E.; Aoki, H.; Yokoyama, T.; Fokt, I.; Hess, K.; Conrad, C.; Madden, T.; Sawaya, R.; et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 2007, 26, 2435–2444. [Google Scholar] [CrossRef] [PubMed]
- Da Fonseca, A.C.C.; Badie, B. Microglia and macrophages in malignant gliomas: Recent discoveries and implications for promising therapies. Clin. Dev. Immunol. 2013, 2013, 264124. [Google Scholar] [CrossRef] [PubMed]
- Badie, B.; Schartner, J.; Prabakaran, S.; Paul, J.; Vorpahl, J. Expression of Fas ligand by microglia: Possible role in glioma immune evasion. J. Neuroimmunol. 2001, 120, 19–24. [Google Scholar] [CrossRef]
- Didenko, V.V.; Ngo, H.N.; Minchew, C.; Baskin, D.S. Apoptosis of T lymphocytes invading glioblastomas multiforme: A possible tumor defense mechanism. J. Neurosurg. 2002, 96, 580–584. [Google Scholar] [CrossRef] [PubMed]
- Ford, A.L.; Foulcher, E.; Lemckert, F.A.; Sedgwick, J.D. Microglia induce CD4 T lymphocyte final effector function and death. J. Exp. Med. 1996, 184, 1737–1745. [Google Scholar] [CrossRef] [PubMed]
- Ebner, F.; Brandt, C.; Thiele, P.; Richter, D.; Schliesser, U.; Siffrin, V.; Schueler, J.; Stubbe, T.; Ellinghaus, A.; Meisel, C.; et al. Microglial activation milieu controls regulatory T cell responses. J. Immunol. 2013, 191, 5594–5602. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Harris, R.A.; Zhang, X.-M. An updated assessment of microglia depletion: Current concepts and future directions. Mol. Brain 2017, 10, 25. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, L.; Ajmone-Cat, M.A.; Cecchetti, S.; Ricci, A.; Bozzuto, G.; Molinari, A.; Manni, I.; Pollo, B.; Scala, S.; Carpinelli, G.; et al. Targeting CXCR4 by a selective peptide antagonist modulates tumor microenvironment and microglia reactivity in a human glioblastoma model. J. Exp. Clin. Cancer Res. 2016, 35, 55. [Google Scholar] [CrossRef] [PubMed]
- Ieranò, C.; Portella, L.; Lusa, S.; Salzano, G.; D’Alterio, C.; Napolitano, M.; Buoncervello, M.; Macchia, D.; Spada, M.; Barbieri, A.; et al. CXCR4-antagonist Peptide R-liposomes for combined therapy against lung metastasis. Nanoscale 2016, 8, 7562–7571. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, A.C.; Romão, L.; Amaral, R.F.; Assad Kahn, S.; Lobo, D.; Martins, S.; de Souza, J.M.; Moura-Neto, V.; Lima, F.R.S. Microglial stress inducible protein 1 promotes proliferation and migration in human glioblastoma cells. Neuroscience 2012, 200, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Cannarile, M.A.; Weisser, M.; Jacob, W.; Jegg, A.-M.; Ries, C.H.; Rüttinger, D. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 2017, 5, 53. [Google Scholar] [CrossRef] [PubMed]
- Butowski, N.; Colman, H.; De Groot, J.F.; Omuro, A.M.; Nayak, L.; Wen, P.Y.; Cloughesy, T.F.; Marimuthu, A.; Haidar, S.; Perry, A.; et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: An Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-Oncology 2016, 18, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Bowman, R.L.; Akkari, L.; Quick, M.L.; Schuhmacher, A.J.; Huse, J.T.; Holland, E.C.; Sutton, J.C.; Joyce, J.A. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science 2016, 352, aad3018. [Google Scholar] [CrossRef] [PubMed]
- Pradel, L.P.; Ooi, C.-H.; Romagnoli, S.; Cannarile, M.A.; Sade, H.; Rüttinger, D.; Ries, C.H. Macrophage Susceptibility to Emactuzumab (RG7155) Treatment. Mol. Cancer Ther. 2016, 15, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Ries, C.H.; Cannarile, M.A.; Hoves, S.; Benz, J.; Wartha, K.; Runza, V.; Rey-Giraud, F.; Pradel, L.P.; Feuerhake, F.; Klaman, I.; et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell 2014, 25, 846–859. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-B.; Zhang, X.; Paul, D.; Kays, L.M.; Gough, W.; Stewart, J.; Uhlik, M.T.; Chen, Q.; Hui, Y.-H.; Zamek-Gliszczynski, M.J.; et al. Identification of LY2510924, a novel cyclic peptide CXCR4 antagonist that exhibits antitumor activities in solid tumor and breast cancer metastatic models. Mol. Cancer Ther. 2015, 14, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, H.; Yang, Y.; Meng, J.; Liu, J.; Wang, C.; Xu, H. A designed peptide targeting CXCR4 displays anti-acute myelocytic leukemia activity in vitro and in vivo. Sci. Rep. 2014, 4, 6610. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ge, Y.; Li, X.; Yang, Y.; Meng, J.; Liu, J.; Wang, C.; Xu, H. Targeting the CXCR4/CXCL12 axis with the peptide antagonist E5 to inhibit breast tumor progression. Signal Transduct. Target. Ther. 2017, 2, 17033. [Google Scholar] [CrossRef] [PubMed]
- Burger, M.; Glodek, A.; Hartmann, T.; Schmitt-Gräff, A.; Silberstein, L.E.; Fujii, N.; Kipps, T.J.; Burger, J.A. Functional expression of CXCR4 (CD184) on small-cell lung cancer cells mediates migration, integrin activation, and adhesion to stromal cells. Oncogene 2003, 22, 8093–8101. [Google Scholar] [CrossRef] [PubMed]
- Portella, L.; Vitale, R.; De Luca, S.; D’Alterio, C.; Ieranò, C.; Napolitano, M.; Riccio, A.; Polimeno, M.N.; Monfregola, L.; Barbieri, A.; et al. Preclinical development of a novel class of CXCR4 antagonist impairing solid tumors growth and metastases. PLoS ONE 2013, 8, e74548. [Google Scholar] [CrossRef] [PubMed]
- Kioi, M.; Vogel, H.; Schultz, G.; Hoffman, R.M.; Harsh, G.R.; Brown, J.M. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Investig. 2010, 120, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-C.; Yu, C.-F.; Hong, J.-H.; Tsai, C.-S.; Chiang, C.-S. Radiation Therapy-Induced Tumor Invasiveness Is Associated with SDF-1-Regulated Macrophage Mobilization and Vasculogenesis. PLoS ONE 2013, 8, e69182. [Google Scholar] [CrossRef] [PubMed]
- Zabel, B.A.; Wang, Y.; Lewén, S.; Berahovich, R.D.; Penfold, M.E.T.; Zhang, P.; Powers, J.; Summers, B.C.; Miao, Z.; Zhao, B.; et al. Elucidation of CXCR7-mediated signaling events and inhibition of CXCR4-mediated tumor cell transendothelial migration by CXCR7 ligands. J. Immunol. 2009, 183, 3204–3211. [Google Scholar] [CrossRef] [PubMed]
- Dillmann, F.; Veldwijk, M.R.; Laufs, S.; Sperandio, M.; Calandra, G.; Wenz, F.; Zeller, W.J.; Fruehauf, S. Plerixafor inhibits chemotaxis toward SDF-1 and CXCR4-mediated stroma contact in a dose-dependent manner resulting in increased susceptibility of BCR-ABL+ cell to Imatinib and Nilotinib. Leuk. Lymphoma 2009, 50, 1676–1686. [Google Scholar] [CrossRef] [PubMed]
- Salacz, M.E.; Kast, R.E.; Saki, N.; Brüning, A.; Karpel-Massler, G.; Halatsch, M.-E. Toward a noncytotoxic glioblastoma therapy: Blocking MCP-1 with the MTZ Regimen. OncoTargets Ther. 2016, 9, 2535–2545. [Google Scholar] [CrossRef] [PubMed]
- Horiguchi, A.; Asano, T.; Kuroda, K.; Sato, A.; Asakuma, J.; Ito, K.; Hayakawa, M.; Sumitomo, M.; Asano, T. STAT3 inhibitor WP1066 as a novel therapeutic agent for renal cell carcinoma. Br. J. Cancer 2010, 102, 1592–1599. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.; Wei, J.; Kong, L.-Y.; Wang, Y.; Priebe, W.; Qiao, W.; Sawaya, R.; Heimberger, A.B. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro-Oncology 2010, 12, 1113–1125. [Google Scholar] [CrossRef] [PubMed]
- Elmore, M.R.P.; Najafi, A.R.; Koike, M.A.; Dagher, N.N.; Spangenberg, E.E.; Rice, R.A.; Kitazawa, M.; Matusow, B.; Nguyen, H.; West, B.L.; et al. Colony-Stimulating Factor 1 Receptor Signaling Is Necessary for Microglia Viability, Unmasking a Microglia Progenitor Cell in the Adult Brain. Neuron 2014, 82, 380–397. [Google Scholar] [CrossRef] [PubMed]
- Strachan, D.C.; Ruffell, B.; Oei, Y.; Bissell, M.J.; Coussens, L.M.; Pryer, N.; Daniel, D. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8(+) T cells. Oncoimmunology 2013, 2, e26968. [Google Scholar] [CrossRef] [PubMed]
- Rios, A.; Hsu, S.H.; Blanco, A.; Buryanek, J.; Day, A.L.; McGuire, M.F.; Brown, R.E. Durable response of glioblastoma to adjuvant therapy consisting of temozolomide and a weekly dose of AMD3100 (plerixafor), a CXCR4 inhibitor, together with lapatinib, metformin and niacinamide. Oncoscience 2016, 3, 156–163. [Google Scholar] [CrossRef] [PubMed]
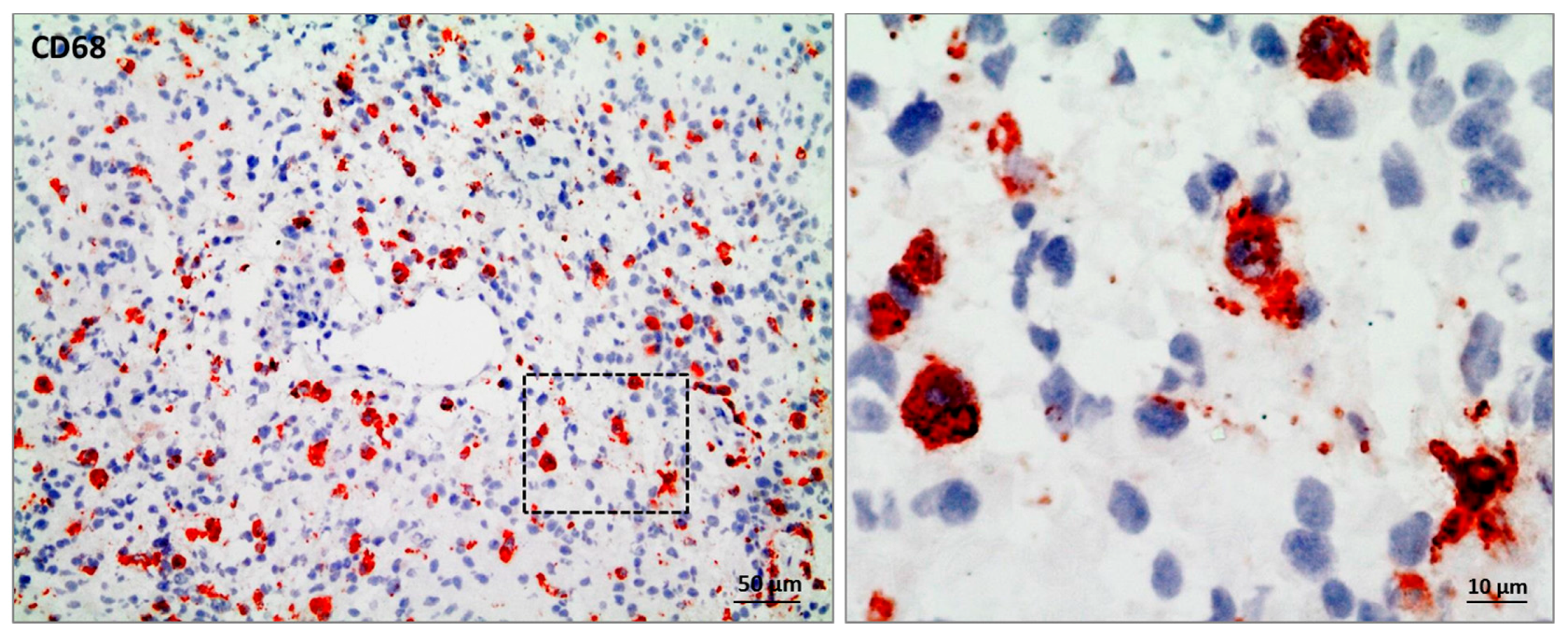
| Marker | Microglia | BMDMs | M1-Like | M2-Like | Species | Reference |
|---|---|---|---|---|---|---|
| CD68 | X | x | human/murine | [7,76] | ||
| IBA1 | X | x | human/murine | [77] | ||
| CD11b | X | x | human/murine | [78,79] | ||
| F4/80 | X | x | murine | [80] | ||
| CX3CR1+/CCR2− | X | human/murine | [81,82] | |||
| CX3CR1−/CCR2+ | x | human/murine | [81,82] | |||
| CD11b+/CD45low | X | human/murine | [7,23] | |||
| CD11b+/CD45high | x | human/murine | [7,23] | |||
| MHC IIhigh | x | human/murine | [83,84] | |||
| CD80high/CD86high | x | human/murine | [84,85] | |||
| CD80low/CD86low | x | human/murine | [84,85] | |||
| CD74 | x | human | [86] | |||
| NF-κB/STAT1 | x | human/murine | [52,56] | |||
| iNOS/NO | x | human/murine | [52,60] | |||
| TMEM119 | X | human | [20,25] | |||
| pSTAT3 | x | human/murine | [73,87] | |||
| CD163 | x | human/murine | [63,73,88] | |||
| CD204 | x | human/murine | [89] | |||
| CD206 | x | human/murine | [65] | |||
| FIZZ1 | (human)/murine | [6,52,90] | ||||
| ARG1 | murine | [6,52,90] |
| Drug Name | Target/Function | Suggested Mode of Action | Study Phase | Tumor Types | Identifier | References |
|---|---|---|---|---|---|---|
| PLX3397 (Pexidartinib) | CSF-1R inhibitor | Reduced GAM infiltration | preclinical | GBM | N/A | [112,164] |
| PLX3397 (Pexidartinib) | CSF-1R inhibitor | GAM elimination | II and I/IIb | rGBM, pGBM | NCT01349036, NCT01790503 | [148] |
| BLZ945 | CSF-1R inhibitor | Inhibition of GAM proliferation, blocking of tumor progression, enhancement of CD8+ T cell infiltration | preclinical | GBM | N/A | [97,165] |
| RG7155 (Emactuzumab) | CSF-1R inhibitor | Alters macrophage polarization and blocks glioma progression | preclinical | GBM | N/A | [97,150] |
| RG7155 (Emactuzumab) | CSF-1R inhibitor | CSF-R1 inhibition | I | GBM | NCT02323191 | [3,149] |
| Plerixafor (AMD3100) | CXCR4 antagonist | Reduced GAM recruitment by inhibition of chemotaxis | I/II | HGG | NCT01977677, NCT01339039 | [144,159,166] |
| Peptide R | CXCR4 antagonist | M1-like polarization | preclinical | GBM | N/A | [144,145] |
| MTZ regimen | CCL2 inhibitor | Reduced GAM recruitment by inhibition of chemotaxis | preclinical | GBM | N/A | [161] |
| WP1066 | STAT3 inhibitor | M1-like polarization through STAT3 blocking | I | GBM, glioma | NCT01904123 | [137,138,163] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roesch, S.; Rapp, C.; Dettling, S.; Herold-Mende, C. When Immune Cells Turn Bad—Tumor-Associated Microglia/Macrophages in Glioma. Int. J. Mol. Sci. 2018, 19, 436. https://doi.org/10.3390/ijms19020436
Roesch S, Rapp C, Dettling S, Herold-Mende C. When Immune Cells Turn Bad—Tumor-Associated Microglia/Macrophages in Glioma. International Journal of Molecular Sciences. 2018; 19(2):436. https://doi.org/10.3390/ijms19020436
Chicago/Turabian StyleRoesch, Saskia, Carmen Rapp, Steffen Dettling, and Christel Herold-Mende. 2018. "When Immune Cells Turn Bad—Tumor-Associated Microglia/Macrophages in Glioma" International Journal of Molecular Sciences 19, no. 2: 436. https://doi.org/10.3390/ijms19020436