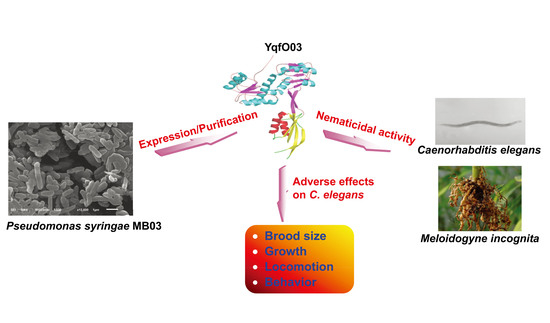
The Nif3-Family Protein YqfO03 from Pseudomonas syringae MB03 Has Multiple Nematicidal Activities against Caenorhabditis elegans and Meloidogyne incognita
Abstract
:1. Introduction
2. Results
2.1. Molecular Characterization and Expression Analysis of the Nif3-family Protein YqfO03
2.2. Nematicidal Activity of Purified YqfO03 against C. elegans and M. incognita
2.3. Effects of Purified YqfO03 on Brood Size and Growth of C. elegans
2.4. Effect of YqfO03 on the Motility of C. elegans
2.5. Effect of YqfO03 on the Behavioral Responses of C. elegans
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains, Plasmids, and Culture Conditions
4.2. Gene Cloning, Protein Expression, and Purification
4.3. Western Blot Analysis
4.4. C. elegans Mortality Assay
4.5. Mortality Assay of M. incognita
4.6. Brood Size Assay and L1 Growth Assay
4.7. Effect of Purified Protein on Nematode Motility
4.8. Behavioral Response of C. elegans
4.9. Microscopy
4.10. Bioinformatics
4.11. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, J.; Liang, L.; Li, J.; Zhang, K.-Q. Nematicidal enzymes from microorganisms and their applications. Appl. Microbiol. Biotechnol. 2013, 97, 7081–7095. [Google Scholar] [CrossRef] [PubMed]
- Abad, P.; Gouzy, J.; Aury, J.M.; Castagnone-Sereno, P.; Danchin, E.G.; Deleury, E.; Perfus-Barbeoch, L.; Anthouard, V.; Artiguenave, F.; Blok, V.C.; et al. Genome sequence of the metazoan plant-parasitic nematode Meloidogyne Incognita. Nat. Biotechnol. 2008, 26, 909–915. [Google Scholar] [CrossRef]
- Trudgill, D.L.; Blok, V.C. Apomictic, polyphagous root-knot nematodes: Exceptionally successful and damaging biotrophic root pathogens. Annu. Rev. Phytopathol. 2001, 39, 53–77. [Google Scholar] [CrossRef]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Kikuchi, T.; Manzanilla-López, R.; Palomares-Rius, J.E.; Wesemael, W.M. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicol, J.M.; Turner, S.J.; Coyne, D.; Den Nijs, L.; Hockland, S.; Maafi, Z.T. Current nematode threats to world agriculture. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J., Gheysen, G., Fenoll, C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 21–43. [Google Scholar]
- Warnock, N.D.; Wilson, L.; Patten, C.; Fleming, C.C.; Maule, A.G.; Dalzell, J.J. Nematode neuropeptides as transgenic nematicides. PLoS Pathogens 2017, 13, e1006237. [Google Scholar] [CrossRef]
- Tian, B.; Yang, J.; Zhang, K.-Q. Bacteria used in the biological control of plant-parasitic nematodes: Populations, mechanisms of action, and future prospects. FEMS Microbiol. Ecol. 2007, 61, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Phani, V.; Shivakumara, T.N.; Davies, K.G.; Rao, U. Meloidogyne incognita fatty acid- and retinol-binding protein (Mi-FAR-1) affects nematode infection of plant roots and the attachment of Pasteuria penetrans endospores. Front. Microbiol. 2017, 8, 2122. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.C.; Zhang, L.; Dai, L.L.; Khan, R.U.; Zou, C.G. mir-67 regulates P. aeruginosa avoidance behavior in C. elegans. Biochem. Biophys. Res. Commun. 2017, 494, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jiang, H.; Cheng, Q.; Chen, J.; Wu, G.; Kumar, A.; Sun, M.; Liu, Z. Enhanced nematicidal potential of the chitinase pachi from Pseudomonas aeruginosa in association with Cry21Aa. Sci. Rep. 2015, 5, 14395. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xiong, J.; Zhou, Q.; Luo, H.; Hu, S.; Xia, L.; Sun, M.; Li, L.; Yu, Z. The diverse nematicidal properties and biocontrol efficacy of Bacillus thuringiensis Cry6A against the root-knot nematode Meloidogyne hapla. J. Invertebr. Pathol. 2015, 125, 73–80. [Google Scholar] [CrossRef]
- Xiong, J.; Zhou, Q.; Luo, H.; Xia, L.; Li, L.; Sun, M.; Yu, Z. Systemic nematicidal activity and biocontrol efficacy of Bacillus firmus against the root-knot nematode Meloidogyne incognita. World J. Microbiol. Biotechnol. 2015, 31, 661–667. [Google Scholar] [CrossRef]
- Kiewnick, S.; Sikora, R.A. Evaluation of Paecilomyces lilacinus strain 251 for the biological control of the northern root-knot nematode Meloidogyne hapla Chitwood. Nematology 2006, 8, 69–78. [Google Scholar]
- Morton, O.C.; Hirsch, P.R.; Kerry, B.R. Infection of plant-parasitic nematodes by nematophagous fungi–A review of the application of molecular biology to understand infection processes and to improve biological control. Nematology 2004, 6, 161–170. [Google Scholar] [CrossRef]
- Tedesco, P.; Di Schiavi, E.; Esposito, F.P.; de Pascale, D. Evaluation of Burkholderia cepacia complex bacteria pathogenicity using Caenorhabditis elegans. Bio. Protoc. 2016, 6, e1964. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.S.; Khan, T.A. Integrated approach for the management of Meloidogyne javanica on eggplant using oil cakes and biocontrol agents. Arch. Phytopathol. Pflanzenschu. 2010, 43, 609–614. [Google Scholar] [CrossRef]
- Terefe, M.; Tefera, T.; Sakhuja, P. Effect of a formulation of Bacillus firmus on root-knot nematode Meloidogyne incognita infestation and the growth of tomato plants in the greenhouse and nursery. J. Invertebr. Pathol. 2009, 100, 94–99. [Google Scholar] [CrossRef]
- Lindeberg, M.; Cunnac, S.; Collmer, A. Pseudomonas syringae type III effector repertoires: Last words in endless arguments. Trends Microbiol. 2012, 20, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Mahajan-Miklos, S.; Tan, M.-W.; Rahme, L.G.; Ausubel, F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa–Caenorhabditis elegans pathogenesis model. Cell 1999, 96, 47–56. [Google Scholar] [CrossRef]
- Zaborin, A.; Romanowski, K.; Gerdes, S.; Holbrook, C.; Lepine, F.; Long, J.; Poroyko, V.; Diggle, S.P.; Wilke, A.; Righetti, K. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. USA 2009, 106, 6327–6332. [Google Scholar] [CrossRef]
- Kirienko, N.V.; Kirienko, D.R.; Larkins-Ford, J.; Wählby, C.; Ruvkun, G.; Ausubel, F.M. Pseudomonas aeruginosa disrupts Caenorhabditis elegans iron homeostasis, causing a hypoxic response and death. Cell Host Microbe 2013, 13, 406–416. [Google Scholar] [CrossRef]
- Lee, D.G.; Urbach, J.M.; Wu, G.; Liberati, N.T.; Feinbaum, R.L.; Miyata, S.; Diggins, L.T.; He, J.; Saucier, M.; Déziel, E. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006, 7, R90. [Google Scholar] [CrossRef] [PubMed]
- Feinbaum, R.L.; Urbach, J.M.; Liberati, N.T.; Djonovic, S.; Adonizio, A.; Carvunis, A.-R.; Ausubel, F.M. Genome-wide identification of Pseudomonas aeruginosa virulence-related genes using a Caenorhabditis elegans infection model. PLoS Pathogens 2012, 8, e1002813. [Google Scholar] [CrossRef] [Green Version]
- Dubern, J.F.; Cigana, C.; De Simone, M.; Lazenby, J.; Juhas, M.; Schwager, S.; Bianconi, I.; Döring, G.; Eberl, L.; Williams, P. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015, 17, 4379–4393. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Sun, Y.; Xie, L.; Yu, H.; Bashir, A.; Li, L. The Pathogenicity of Pseudomonas syringae MB03 against Caenorhabditis elegans and the transcriptional response of nematicidal genes upon different nutritional conditions. Front. Microbiol. 2016, 7, 805. [Google Scholar] [CrossRef] [PubMed]
- Kaletta, T.; Hengartner, M.O. Finding function in novel targets: C. elegans as a model organism. Nat. Rev. Drug Discov. 2006, 5, 387–398. [Google Scholar] [CrossRef]
- Galperin, M.Y.; Koonin, E.V. “Conserved hypothetical” proteins: Prioritization of targets for experimental study. Nucleic Acids Res. 2004, 32, 5452–5463. [Google Scholar] [CrossRef] [PubMed]
- Baysal, Ö.; Lai, D.; Xu, H.-H.; Siragusa, M.; Çalışkan, M.; Carimi, F.; Da Silva, J.A.T.; Tör, M. A proteomic approach provides new insights into the control of soil-borne plant pathogens by Bacillus species. PLoS ONE 2013, 8, e53182. [Google Scholar] [CrossRef]
- Gupta, A.; Kapil, R.; Dhakan, D.B.; Sharma, V.K. MP3: A software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PLoS ONE 2014, 9, e93907. [Google Scholar] [CrossRef] [PubMed]
- Godsey, M.H.; Minasov, G.; Shuvalova, L.; Brunzelle, J.S.; Vorontsov, I.I.; Collart, F.R.; Anderson, W.F. The 2.2 Å resolution crystal structure of Bacillus cereus Nif3-family protein YqfO reveals a conserved dimetal-binding motif and a regulatory domain. Protein Sci. 2007, 16, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Ladner, J.E.; Obmolova, G.; Teplyakov, A.; Howard, A.J.; Khil, P.P.; Camerini-Otero, R.D.; Gilliland, G.L. Crystal structure of Escherichia coli protein YbgI, a toroidal structure with a dinuclear metal site. BMC Struct. Biol. 2003, 3, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saikatendu, K.S.; Zhang, X.; Kinch, L.; Leybourne, M.; Grishin, N.V.; Zhang, H. Structure of a conserved hypothetical protein SA1388 from S. aureus reveals a capped hexameric toroid with two PII domain lids and a dinuclear metal center. BMC Struct. Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Tomoike, F.; Wakamatsu, T.; Nakagawa, N.; Kuramitsu, S.; Masui, R. Crystal structure of the conserved hypothetical protein TTHA1606 from Thermus thermophilus HB8. Proteins 2009, 76, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Auld, D.S. Zinc coordination sphere in biochemical zinc sites. In Zinc Biochemistry, Physiology, and Homeostasis; Maret, W., Ed.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2001; pp. 85–127. [Google Scholar]
- Cahan, R.; Axelrad, I.; Safrin, M.; Ohman, D.E.; Kessler, E. A secreted aminopeptidase of Pseudomonas aeruginosa identification, primary structure, and relationship to other aminopeptidases. J. Biol. Chem. 2001, 276, 43645–43652. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Xiong, J.; Zhou, Q.; Xia, L.; Yu, Z. The effects of Bacillus thuringiensis Cry6A on the survival, growth, reproduction, locomotion, and behavioral response of Caenorhabditis elegans. Appl. Microbiol. Biotechnol. 2013, 97, 10135–10142. [Google Scholar] [CrossRef]
- Li, Q.; Yan, Q.; Chen, J.; He, Y.; Wang, J.; Zhang, H.; Yu, Z.; Li, L. Molecular characterization of an ice nucleation protein variant (InaQ) from Pseudomonas syringae and the analysis of its transmembrane transport activity in Escherichia coli. Int. J. Biol. Sci. 2012, 8, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011. [Google Scholar]
- Bischof, L.J.; Huffman, D.L.; Aroian, R.V. Assays for toxicity studies in C. elegans with Bt crystal proteins. Methods Mol. Biol. 2006, 351, 139–154. [Google Scholar] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Wei, J.-Z.; Hale, K.; Carta, L.; Platzer, E.; Wong, C.; Fang, S.-C.; Aroian, R.V. Bacillus thuringiensis crystal proteins that target nematodes. Proc. Natl. Acad. Sci. USA 2003, 100, 2760–2765. [Google Scholar] [CrossRef]
- Tavernarakis, N.; Shreffler, W.; Wang, S.; Driscoll, M. unc-8, a DEG/ENaC family member, encodes a subunit of a candidate mechanically gated channel that modulates C. elegans locomotion. Neuron 1997, 18, 107–119. [Google Scholar] [CrossRef]
- Hasshoff, M.; Böhnisch, C.; Tonn, D.; Hasert, B.; Schulenburg, H. The role of Caenorhabditis elegans insulin-like signaling in the behavioral avoidance of pathogenic Bacillus thuringiensis. FASEB J. 2007, 21, 1801–1812. [Google Scholar] [CrossRef]








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manan, A.; Bazai, Z.A.; Fan, J.; Yu, H.; Li, L. The Nif3-Family Protein YqfO03 from Pseudomonas syringae MB03 Has Multiple Nematicidal Activities against Caenorhabditis elegans and Meloidogyne incognita. Int. J. Mol. Sci. 2018, 19, 3915. https://doi.org/10.3390/ijms19123915
Manan A, Bazai ZA, Fan J, Yu H, Li L. The Nif3-Family Protein YqfO03 from Pseudomonas syringae MB03 Has Multiple Nematicidal Activities against Caenorhabditis elegans and Meloidogyne incognita. International Journal of Molecular Sciences. 2018; 19(12):3915. https://doi.org/10.3390/ijms19123915
Chicago/Turabian StyleManan, Abdul, Zahoor Ahmad Bazai, Jin Fan, Huafu Yu, and Lin Li. 2018. "The Nif3-Family Protein YqfO03 from Pseudomonas syringae MB03 Has Multiple Nematicidal Activities against Caenorhabditis elegans and Meloidogyne incognita" International Journal of Molecular Sciences 19, no. 12: 3915. https://doi.org/10.3390/ijms19123915