δ-Tocotrienol, Isolated from Rice Bran, Exerts an Anti-Inflammatory Effect via MAPKs and PPARs Signaling Pathways in Lipopolysaccharide-Stimulated Macrophages
Abstract
:1. Introduction
2. Results
2.1. Isolation and Purification of δ-Tocotrienol from Rice Bran
2.2. The Toxicity of δ-Tocotrienol on RAW264.7 Cells
2.3. δ-Tocotrienol Downregulated the Expressions of Proinflammatory Factors
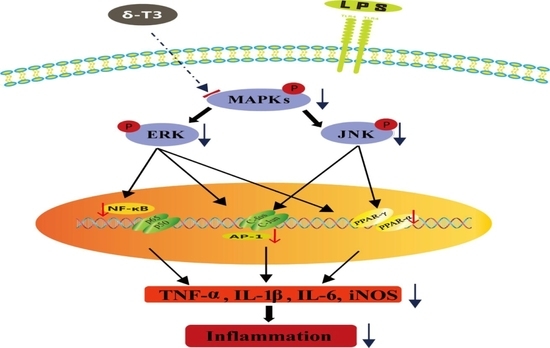
2.4. Effect of δ-Tocotrienol on MAPKs in LPS-Stimulated RAW264.7 Cells
2.5. Effect of δ-Tocotrienol on NF-κB /AP-1 Activities and Translocations
2.6. Effect of δ-Tocotrienol on the Activities of PPARα and PPARγ In Vitro Models
2.7. Effect of δ-Tocotrienol on MAPKs and PPARs Signaling Models
3. Discussion
4. Materials and Methods
4.1. Materials and Reagents
4.2. Isolation of Rice Bran Oil by Supercritical Carbon Dioxide Extraction
4.3. High Performance Liquid Chromatography (HPLC)
4.4. Cell Culture
4.5. Cell Viability Assay
4.6. Observation of Morphological Changes
4.7. RNA Isolation and RT-qPCR
4.8. Extraction of Nuclear and Cytosolic Proteins
4.9. Western Blot Analysis
4.10. Luciferase Reporter Assay
4.11. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Shao, J.; Li, Y.; Wang, Z.; Xiao, M.; Yin, P.; Lu, Y.; Qian, X.; Xu, Y.; Liu, J. 7b, a novel naphthalimide derivative, exhibited anti-inflammatory effects via targeted-inhibiting TAK1 following down-regulation of ERK1/2-and p38 MAPK-mediated activation of NF-κB in LPS-stimulated RAW264.7 macrophages. Int. Immunopharmacol. 2013, 17, 216–228. [Google Scholar] [CrossRef] [PubMed]
- Nahar, P.P.; Driscoll, M.V.; Li, L.; Slitt, A.L.; Seeram, N.P. Phenolic mediated anti-inflammatory properties of a maple syrup extract in RAW 264.7 murine macrophages. J. Funct. Foods 2014, 6, 126–136. [Google Scholar] [CrossRef]
- Kim, K.S.; Lee, D.S.; Bae, G.S.; Park, S.J.; Kang, D.G.; Lee, H.S.; Oh, H.; Kim, Y.C. The inhibition of JNK MAPK and NF-κB signaling by tenuifoliside A isolated from Polygala tenuifolia in lipopolysaccharide-induced macrophages is associated with its anti-inflammatory effect. Eur. J. Pharmacol. 2013, 721, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.C.; Cho, W.K.; Jeong, Y.H.; Im, G.Y.; Lee, K.J.; Yang, H.J.; Ma, J.Y. Anti-inflammatory effect of Sosihotang via inhibition of nuclear factor-κB and mitogen-activated protein kinases signaling pathways in lipopolysaccharide- stimulated RAW264.7 macrophage cells. Food Chem. Toxicol. 2013, 53, 343–351. [Google Scholar] [CrossRef] [PubMed]
- An, H.J.; Kim, I.T.; Park, H.J.; Kim, H.M.; Choi, J.H.; Lee, K.T. Tormentic acid, a triterpenoid saponin, isolated from Rosa rugosa, inhibited LPS-induced iNOS, COX-2, and TNF-α expression through inactivation of the nuclear factor-κB pathway in RAW 264.7 macrophages. Int. Immunopharmacol. 2011, 11, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. n-3 polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am. J. Clin. Nutr. 2006, 83, 77–109. [Google Scholar] [CrossRef] [PubMed]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [CrossRef] [PubMed]
- Utreras, E.; Futatsugi, A.; Rudrabhatla, P.; Keller, J.; Iadarola, M.J.; Pant, H.C.; Kulkarni, A.B. Tumor necrosis factor-α regulates cyclin-dependent kinase 5 activity during pain signaling through transcriptional activation of p35. J. Biol. Chem. 2009, 284, 2275–2284. [Google Scholar] [CrossRef] [PubMed]
- Qiang, L.; Wang, L. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparγ. Cell 2012, 150, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Barish, G.D.; Narkar, V.A.; Evans, R.M. PPARδ: A dagger in the heart of the metabolic syndrome. J. Clin. Investig. 2006, 116, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.; Kang, Z.; Wong, C. Vitamin E tocotrienols improve insulin sensitivity through activating peroxisome proliferator-activated receptors. Mol. Nutr. Food Res. 2010, 54, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, L.; Siersbaek, M.; Mandrup, S. PPARs: Fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 2012, 23, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Soccio, R.E.; Chen, E.R.; Lazar, M.A. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014, 20, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Reis, J.C.; Papasian, C.J.; Morrison, D.C.; Qureshi, N. Tocotrienols inhibit lipopolysaccharide-induced pro-inflammatory cytokines in macrophages of female mice. Lipids Health Dis. 2010, 9, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Tan, X.; Reis, J.C.; Badr, M.Z.; Papasian, C.J. Inhibition of nitric oxide in LPS-stimulated macrophages of young and senescent mice by δ-tocotrienol and quercetin. Lipids Health Dis. 2011, 10, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Laborte, A.G.; Gutierrez, M.A.; Balanza, J.G.; Saito, K.; Zwart, S.J.; Boschetti, M.; Murty, M.V.R.; Villano, L.; Aunario, J.K.; Reinke, R.; et al. RiceAtlas, a spatial database of global rice calendars and production. Sci. Data 2017, 4, 170074–170083. [Google Scholar] [CrossRef] [PubMed]
- Park, H.Y.; Lee, K.W.; Choi, H. Rice bran constituents: Immunomodulatory and therapeutic activities. Food Funct. 2017, 8, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.T. Rice: Origin, Domestication, and Diversification. 1. In Rice: Origin, History, Technology, and Production; Smith, C.W., Dilday, R., Eds.; Horboken: Chichester, NJ, USA, 2003; pp. 3–25. [Google Scholar]
- Evans, H.M.; Bishop, K.S. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science 1922, 56, 650–651. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, A.A.; Mo, H.; Packer, L.; Peterson, D.M. Isolation and identification of novel tocotrienol from rice bran with hypocholesterolemic, antioxidant, and antitumor properties. J. Agric. Food Chem. 2000, 48, 3130–3140. [Google Scholar] [CrossRef] [PubMed]
- Tuncel, N.B.; Yılmaz, N. δ-Tocotrienol content, phenolic acid profiles and antioxidant activity of rice milling fractions. Eur. Food Res. Technol. 2011, 233, 577–585. [Google Scholar] [CrossRef]
- Xu, W.L.; Liu, J.R.; Liu, H.K.; Qi, G.Y.; Sun, X.R.; Sun, W.G.; Chen, B.Q. Inhibition of proliferation and induction of apoptosis by γ-tocotrienol in human colon carcinoma HT-29 cells. Nutrition 2009, 25, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Brzuskiewicz, L. Separation of tocopherol and tocotrienol isomers using normal- and reverse-phase liquid chromatography. Anal. Biochem. 1989, 180, 368–373. [Google Scholar] [CrossRef]
- Mustacich, D.J.; Leonard, S.W.; Patel, N.K.; Traber, M.G. α-Tocopherol β-oxidation localized to rat liver mitochondria. Free Radic. Biol. Med. 2010, 48, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, H.; Ahad, A.; Iqbal, J.; Siddiqui, W.A. Pharmacological potential of tocotrienols: A review. Nutr. Metab. 2014, 11, 52–73. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.; Tao, H.; Metrione, M.; Hajri, T. Very low density lipoprotein receptor expression is a determinant factor in adipose tissue inflammation and adipocyte-macrophage interaction. J. Biol. Chem. 2013, 289, 1688–1703. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lin, Q.; Yang, T.; Zeng, L.; Shi, L.; Chen, Y.; Luo, F. Oat β-glucan prevents DSS induced colitis in mice by (or through) decreasing the expression of inflammatory cytokines TNF-α, IL-1β, IL-6 and iNOS. Food Funct. 2015, 11, 3454–3463. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.W.; Pyo, Y.G.; Lee, J.; Lee, J.S.; Kim, B.H.; Kim, I.H. Concentrations of tocols and γ-oryzanol compounds in rice bran oil obtained by fractional extraction with supercritical carbon dioxide. J. Oleo Sci. 2014, 63, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Guha, M.; Mackman, N. LPS induction of gene expression in human monocytes. Cell Signal. 2001, 13, 85–94. [Google Scholar] [CrossRef]
- Dali-Youcef, N.; Ricci, R. Signalling networks governing metabolic inflammation. Handb. Exp. Pharmacol. 2016, 233, 195–220. [Google Scholar] [PubMed]
- Guo, T.; Lin, Q.; Li, X.; Nie, Y.; Wang, L.; Shi, L.; Xu, W.; Hu, T.; Guo, T.; Luo, F. Octacosanol attenuates inflammation in both RAW264.7 macrophages and a mouse model of colitis. J. Agric. Food Chem. 2017, 65, 3647–3658. [Google Scholar] [CrossRef] [PubMed]
- Kaimoto, S.; Hoshino, A.; Ariyoshi, M. Activation of PPARα in the early stage of heart failure maintained myocardial function and energetics in pressure overload heart failure. Am. J. Physiol. Heart Circ. Physiol. 2016, 312, H305–H313. [Google Scholar] [CrossRef] [PubMed]
- Ajith, T.A.; Jayakumar, T.G. Peroxisome proliferator-activated receptors in cardiac energy metabolism and cardiovascular disease. Clin. Exp. Pharmacol. Physiol. 2016, 43, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; Degirolamo, C.; Mariani-Costantini, R.; Palasciano, G.; Moschetta, A. Lipid-sensing nuclear receptors in the pathophysiology and treatment of the metabolic syndrome. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011, 5, 562–587. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.; Henry, R.R. Thiazolidinedione safety. Expert Opin. Drug Saf. 2012, 11, 565–579. [Google Scholar] [PubMed]
- Aggarwal, B.B.; Sundaram, C.; Prasad, S.; Kannappan, R. Tocotrienols, the vitamin E of the 21st century: Its potential against cancer and other chronic diseases. Biochem. Pharmacol. 2010, 80, 1613–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesaretnam, K.; Devasagayam, T.P.A.; Singh, B.B.; Basiron, Y. Influence of palm oil or its tocotrienol-rich fraction on the lipid peroxidation potential of rat liver mitochondria and microsomes. Biochem. Mol. Biol. Int. 1993, 30, 159–167. [Google Scholar] [PubMed]
- Serbinova, B.; Kagan, Y.; Han, D.; Packer, L. Free radical recycling and intramembrane mobility in the antioxidant properties of α-tocopherol and α-tocotrienol. Free Radic. Biol. Med. 1991, 10, 263–275. [Google Scholar] [CrossRef]
- Ng, L.T.; Ko, HJ. Comparative effects of tocotrienol-rich fraction, α-tocopherol and α-tocopheryl acetate on inflammatory mediators and nuclear factor κB expression in mouse peritoneal macrophages. Food Chem. 2012, 134, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q. Natural forms of vitamin E: Metabolism, antioxidant, and anti-inflammatory activities and their role in disease prevention and therapy. Free Radic. Biol. Med. 2014, 72, 76–90. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Elson-Schwab, I.; Courtemanche, C.; Ames, B.N. γ-Tocopherol and its major metabolite, in contrast to α-tocopherol, inhibit cycloox-ygenase activity in macrophages and epithelial cells. Proc. Natl. Acad. Sci. USA 2000, 97, 11494–11499. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Yin, X.; Lill, M.A.; Danielson, M.L.; Freiser, H.; Huang, J. Long-chain carboxychromanols metabolites of vitamin E are potent inhibitors of cyclooxygenases. Proc. Natl. Acad. Sci. USA 2008, 105, 20464–20469. [Google Scholar] [CrossRef] [PubMed]
- Montagnani, M.; Marzagalli, M.; Moretti, R.M. δ-Tocotrienol triggers endoplasmic reticulum stress mediated apoptosis in human melanoma cells. Sci. Rep. 2016, 6, 30502–30515. [Google Scholar] [CrossRef] [PubMed]
- Michalik, L.; Auwerx, J.; Berger, J.P.; Chatterjee, V.K.; Glass, C.K.; Gonzalez, F.J.; Grimaldi, P.A.; Kadowaki, T.; Lazar, M.A.; O’Rahilly, S.; et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol. Rev. 2006, 58, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Delerive, P.; De Bosscher, K.; Vanden Berghe, W.; Fruchart, J.C.; Haegeman, G.; Staels, B. DNA binding-independent induction of IκBα gene transcription by PPARα. Mol. Endocrinol. 2002, 16, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Ahmadian, M.; Suh, J.M.; Hah, N. PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 2013, 19, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Red Eagle, A.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARγ controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hevener, A.L.; Olefsky, J.M.; Reichart, D.; Nguyen, M.T.; Bandyopadyhay, G.; Leung, H.Y.; Watt, M.J.; Benner, C.; Febbraio, M.A.; Nguyen, A.K.; Folian, B.; Subramaniam, S.; et al. Macrophage PPARα is required for normal skeletal muscle and hepatic insulin sensitivity and full antidiabetic effects of thiazolidinediones. J. Clin. Investig. 2007, 117, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Regulation of nutrient metabolism and inflammation. Results Probl. Cell Differ. 2010, 52, 13–25. [Google Scholar] [PubMed]
- Necela, B.M.; Su, W.; Thompson, E.A. Toll-like receptor 4 mediates cross-talk between peroxisome proliferator-activated receptor α and nuclear factor-κB in macrophages. Immunology 2008, 125, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Neve, B.P.; Fruchart, J.C.; Staels, B. Role of the peroxisome proliferator-activated receptors (PPAR) in atherosclerosis. Biochem. Pharmacol. 2000, 60, 1245–1250. [Google Scholar] [CrossRef]
- Tornatore, L.; Thotakura, A.K.; Bennett, J.; Moretti, M.; Franzoso, G. The nuclear factor κB signaling pathway: Integrating metabolism with inflammation. Trends Cell Biol. 2012, 22, 557–566. [Google Scholar] [CrossRef] [PubMed]









© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, J.; Yang, T.; Xu, Y.; Luo, Y.; Zhong, X.; Shi, L.; Hu, T.; Guo, T.; Nie, Y.; Luo, F.; et al. δ-Tocotrienol, Isolated from Rice Bran, Exerts an Anti-Inflammatory Effect via MAPKs and PPARs Signaling Pathways in Lipopolysaccharide-Stimulated Macrophages. Int. J. Mol. Sci. 2018, 19, 3022. https://doi.org/10.3390/ijms19103022
Shen J, Yang T, Xu Y, Luo Y, Zhong X, Shi L, Hu T, Guo T, Nie Y, Luo F, et al. δ-Tocotrienol, Isolated from Rice Bran, Exerts an Anti-Inflammatory Effect via MAPKs and PPARs Signaling Pathways in Lipopolysaccharide-Stimulated Macrophages. International Journal of Molecular Sciences. 2018; 19(10):3022. https://doi.org/10.3390/ijms19103022
Chicago/Turabian StyleShen, Junjun, Tao Yang, Youzhi Xu, Yi Luo, Xinyue Zhong, Limin Shi, Tao Hu, Tianyi Guo, Ying Nie, Feijun Luo, and et al. 2018. "δ-Tocotrienol, Isolated from Rice Bran, Exerts an Anti-Inflammatory Effect via MAPKs and PPARs Signaling Pathways in Lipopolysaccharide-Stimulated Macrophages" International Journal of Molecular Sciences 19, no. 10: 3022. https://doi.org/10.3390/ijms19103022