Functional Analyses of RUNX3 and CaMKIINα in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINα and Dichotomous Roles for RUNX3 Transcript Variants
Abstract
:1. Introduction
2. Results
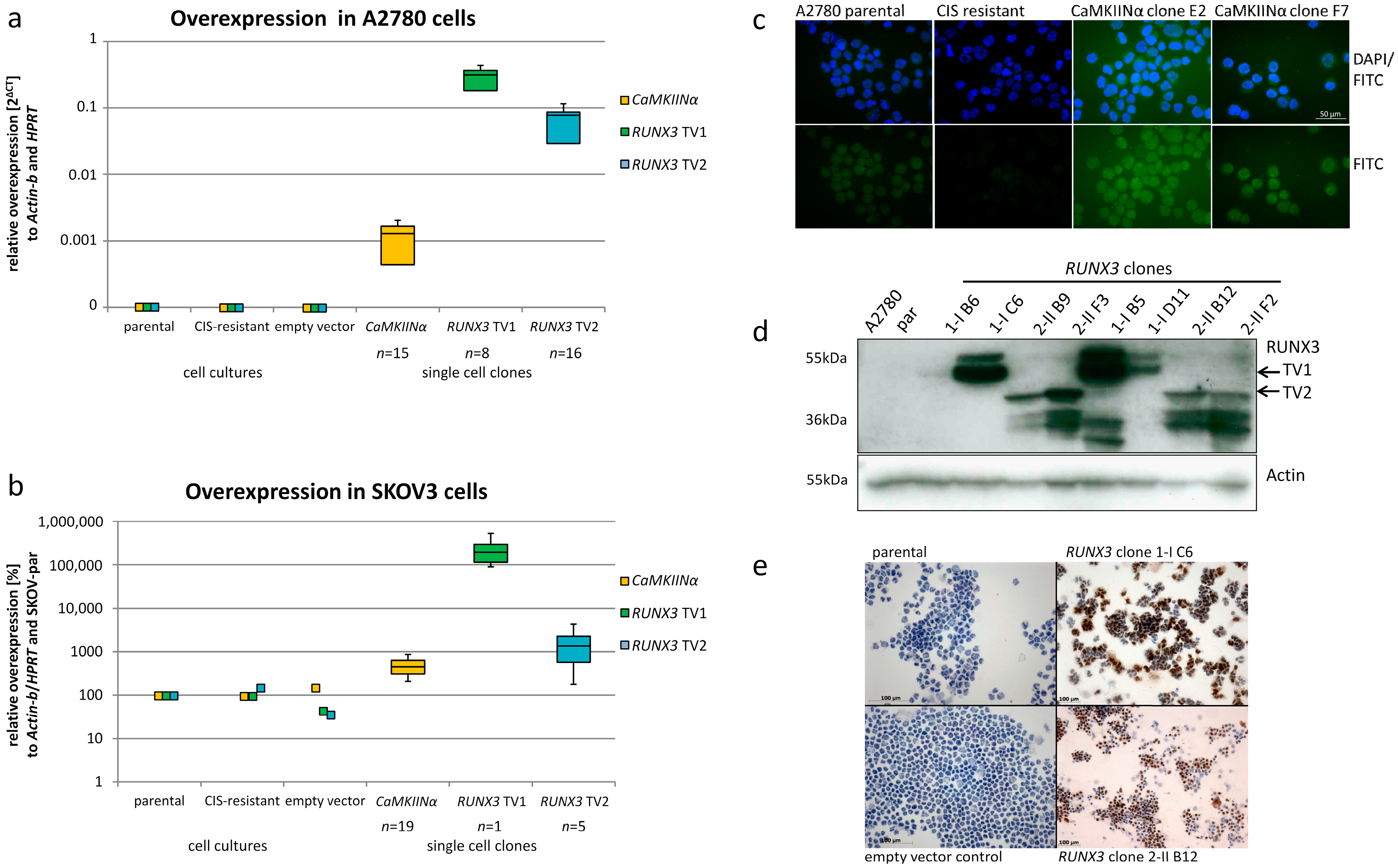
2.1. Ectopic Expression of CaMKIINα and RUNX3 in EOC Cells
2.2. Cisplatin Sensitivity in CaMKIINα Overexpressing Resistant Cells
2.3. Cisplatin Sensitivity under the Influence of RUNX3
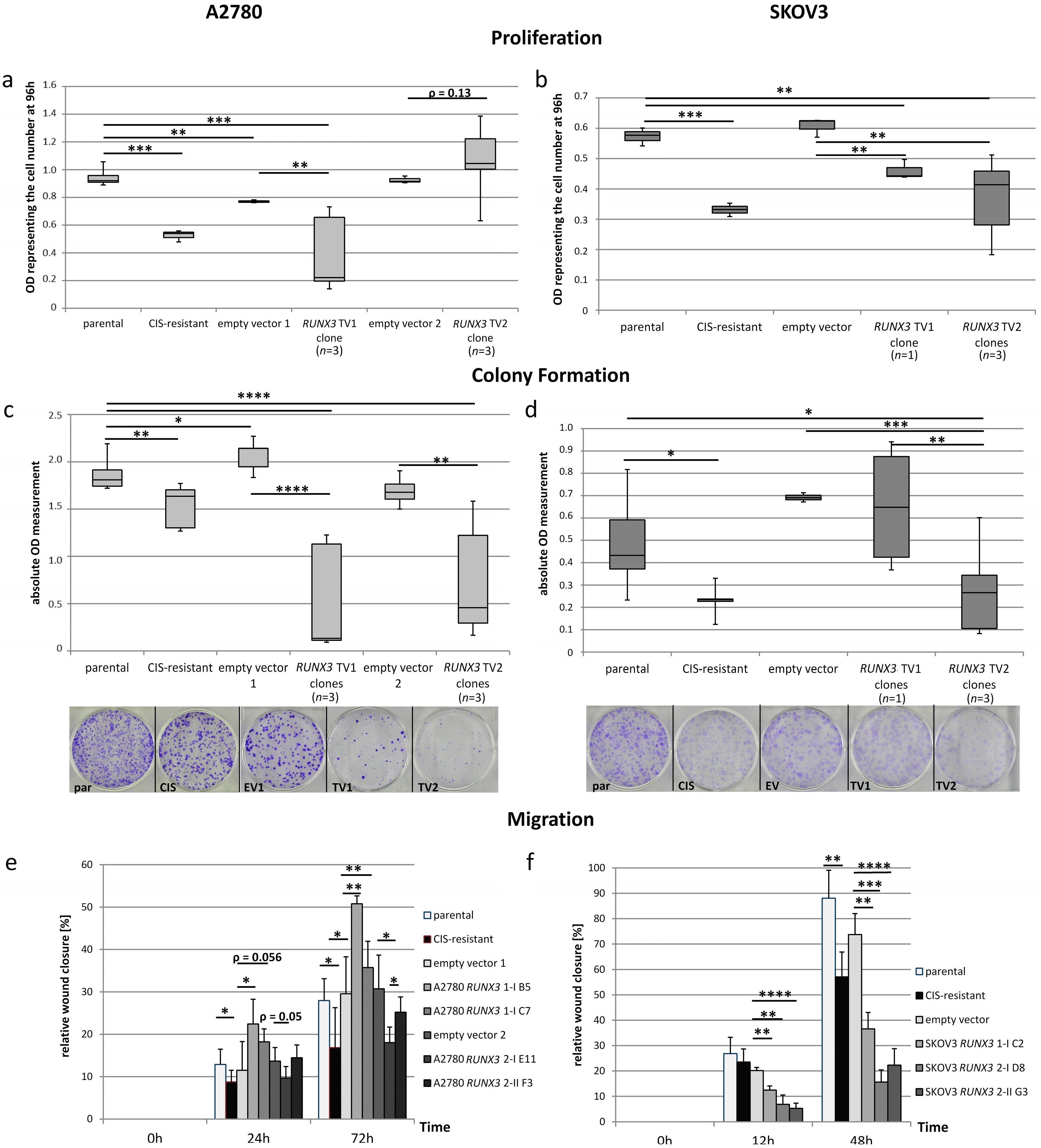
2.4. Reduction of Cellular Functions through CaMKIINα Overexpression
2.5. Dualistic Effect of the RUNX3 Transcript Variants on Cellular Functions
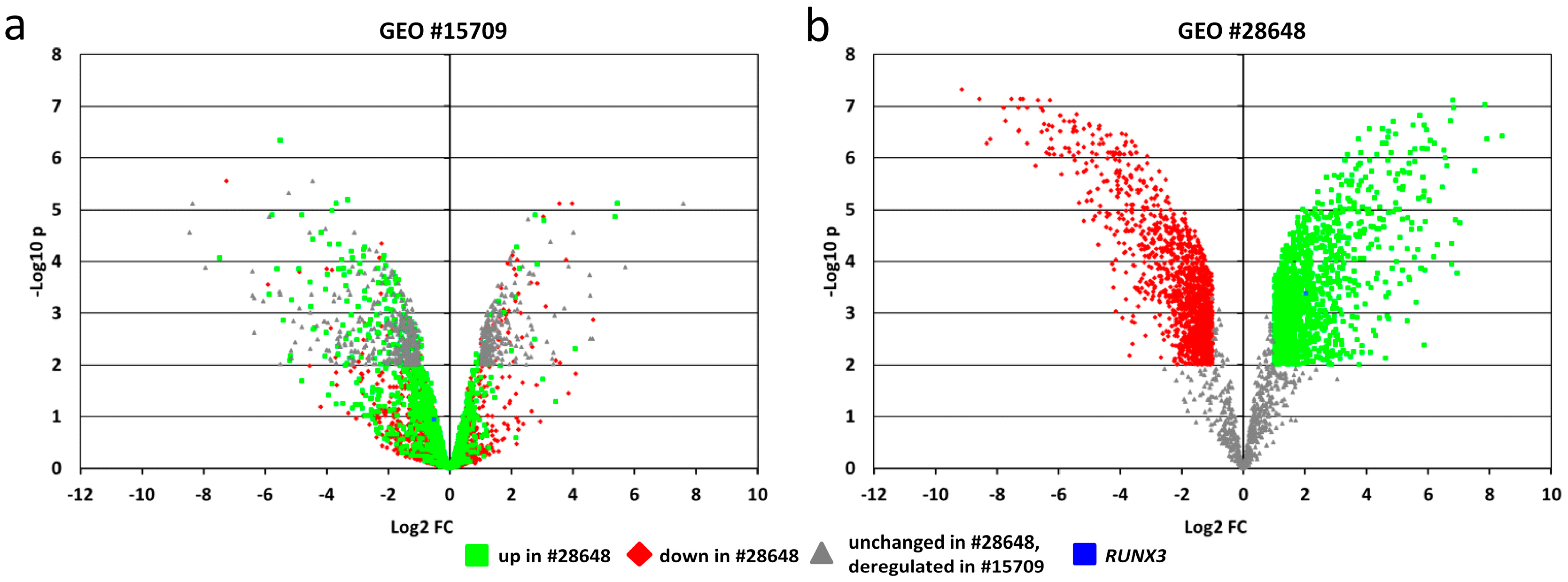
2.6. In-Silico Analysis of Published Datasets for RUNX3
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. Transfection and Single Cell Clone Generation
4.3. Immunocytochemical Staining Assays
4.4. Gene expression Analysis
4.5. Western Blot Analysis
4.6. IC50 Determination Against Cisplatin
4.7. Cell Proliferation Assay
4.8. Colony Formation Assay
4.9. Migration-Scratch Assay
4.10. Flow Cytometry
4.11. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| EOC | epithelial ovarian cancer |
| IC | inhibitory concentration |
| TV | transcript variant |
| 5yr-SR | 5-year-survival-rate |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef] [PubMed]
- Robert-Koch-Institut (RKI). Krebs in Deutschland 2011/2012; Robert-Koch-Institut und GEKID: Berlin, Germany, 2015. [Google Scholar]
- Vaughan, S.; Coward, J.I.; Bast, R.C., Jr.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D.; et al. Rethinking ovarian cancer: Recommendations for improving outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [CrossRef] [PubMed]
- National-Cancer-Institute SEER. SEER Cancer Statistics Review, 1975–2012; SEER: Bethesda, MD, USA, 2015. [Google Scholar]
- McCluggage, W.G. Morphological subtypes of ovarian carcinoma: A review with emphasis on new developments and pathogenesis. Pathology 2011, 43, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Greenlee, R.T.; Hill-Harmon, M.B.; Murray, T.; Thun, M. Cancer statistics, 2001. CA Cancer J. Clin. 2001, 51, 15–36. [Google Scholar] [CrossRef] [PubMed]
- International Agency for Research on Cancer (IARC). GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012; IARC: Lyon, France, 2012. [Google Scholar]
- McGuire, W.P. Maintenance therapy for ovarian cancer: Of Helsinki and Hippocrates. J. Clin. Oncol. 2009, 27, 4633–4634. [Google Scholar] [CrossRef] [PubMed]
- Omura, G.; Blessing, J.A.; Ehrlich, C.E.; Miller, A.; Yordan, E.; Creasman, W.T.; Homesley, H.D. A randomized trial of cyclophosphamide and doxorubicin with or without cisplatin in advanced ovarian carcinoma. A Gynecologic Oncology Group Study. Cancer 1986, 57, 1725–1730. [Google Scholar] [CrossRef]
- Coleman, M.P.; Forman, D.; Bryant, H.; Butler, J.; Rachet, B.; Maringe, C.; Nur, U.; Tracey, E.; Coory, M.; Hatcher, J.; et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): An analysis of population-based cancer registry data. Lancet 2011, 377, 127–138. [Google Scholar] [CrossRef]
- Bijron, J.G.; Bol, G.M.; Verheijen, R.H.; van Diest, P.J. Epigenetic biomarkers in the diagnosis of ovarian cancer. Expert Opin. Med. Diagn. 2012, 6, 421–438. [Google Scholar] [CrossRef] [PubMed]
- Barton, C.A.; Hacker, N.F.; Clark, S.J.; O’Brien, P.M. DNA methylation changes in ovarian cancer: Implications for early diagnosis, prognosis and treatment. Gynecol. Oncol. 2008, 109, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Hafner, N.; Steinbach, D.; Jansen, L.; Diebolder, H.; Durst, M.; Runnebaum, I.B. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int. J. Cancer 2016, 138, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.H.; Mukherji, S.; Soderling, T.R. Characterization of a calmodulin kinase II inhibitor protein in brain. Proc. Natl. Acad. Sci. USA 1998, 95, 10890–10895. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.H.; Mukherji, S.; Soderling, T.R. Calcium/calmodulin-dependent protein kinase II inhibitor protein: Localization of isoforms in rat brain. Neuroscience 2001, 102, 767–777. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Z.; Guo, S.; Wu, L.; Li, M.; Yang, J.; Chen, R.; Xu, H.; Cai, S.; Chen, H.; et al. The tumor suppressive role of CAMK2N1 in castration-resistant prostate cancer. Oncotarget 2014, 5, 3611–3621. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Yang, Y.; Wang, C.; Hui, N.; Gu, L.; Zhong, H.; Cai, Z.; Wang, Q.; Zhang, Q.; Li, N.; et al. Endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest and apoptosis through down-regulation of the phosphatidylinositide 3-kinase/Akt/HDM2 pathway. J. Biol. Chem. 2009, 284, 24773–24782. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.; Salzano, M.; De Falco, V.; Mian, C.; Barollo, S.; Secondo, A.; Bifulco, M.; Vitale, M. Calcium/Calmodulin-dependent protein kinase II and its endogenous inhibitor alpha in medullary thyroid cancer. Clin. Cancer Res. 2014, 20, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Chuang, L.S.; Ito, Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 2010, 29, 2605–2615. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Ju, H.; Kim, M.; Kang, C. Increased genetic susceptibility to intestinal-type gastric cancer is associated with increased activity of the RUNX3 distal promoter. Cancer 2011, 117, 5161–5171. [Google Scholar] [CrossRef] [PubMed]
- Egawa, T.; Tillman, R.E.; Naoe, Y.; Taniuchi, I.; Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007, 204, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Chuang, L.S.; Kimura, S.; Lai, S.K.; Ong, C.W.; Yan, B.; Salto-Tellez, M.; Choolani, M.; Ito, Y. RUNX3 functions as an oncogene in ovarian cancer. Gynecol. Oncol. 2011, 122, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Bae, S.C.; Chuang, L.S. The RUNX family: Developmental regulators in cancer. Nat. Rev. Cancer 2015, 15, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Barghout, S.H.; Zepeda, N.; Vincent, K.; Azad, A.K.; Xu, Z.; Yang, C.; Steed, H.; Postovit, L.M.; Fu, Y. RUNX3 contributes to carboplatin resistance in epithelial ovarian cancer cells. Gynecol. Oncol. 2015, 138, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Lanczky, A.; Szallasi, Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer 2012, 19, 197–208. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Balch, C.; Montgomery, J.S.; Jeong, M.; Chung, J.H.; Yan, P.; Huang, T.H.; Kim, S.; Nephew, K.P. Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med. Genom. 2009, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Zeller, C.; Dai, W.; Steele, N.L.; Siddiq, A.; Walley, A.J.; Wilhelm-Benartzi, C.S.; Rizzo, S.; van der Zee, A.; Plumb, J.A.; Brown, R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 2012, 31, 4567–4576. [Google Scholar] [CrossRef] [PubMed]
- Bachvarov, D.; L’Esperance, S.; Popa, I.; Bachvarova, M.; Plante, M.; Tetu, B. Gene expression patterns of chemoresistant and chemosensitive serous epithelial ovarian tumors with possible predictive value in response to initial chemotherapy. Int. J. Oncol. 2006, 29, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Lim, A.C.; Salto-Tellez, M.; Motoda, L.; Osato, M.; Chuang, L.S.; Lee, C.W.; Voon, D.C.; Koo, J.K.; Wang, H.; et al. RUNX3 attenuates beta-catenin/T cell factors in intestinal tumorigenesis. Cancer Cell 2008, 14, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, Y.; Miyauchi, M.; Kizawa, I.; Oomori, K.; Kato, K. Establishment of a cisplatin-resistant human ovarian cancer cell line. J. Natl. Cancer Inst. 1986, 77, 1181–1185. [Google Scholar] [PubMed]
- Perez, R.P.; Handel, L.M.; Hamilton, T.C. Potentiation of cisplatin cytotoxicity in human ovarian carcinoma cell lines by trifluoperazine, a calmodulin inhibitor. Gynecol. Oncol. 1992, 46, 82–87. [Google Scholar] [CrossRef]
- Zhang, J.; Li, N.; Yu, J.; Zhang, W.; Cao, X. Molecular cloning and characterization of a novel calcium/calmodulin-dependent protein kinase II inhibitor from human dendritic cells. Biochem. Biophys. Res. Commun. 2001, 285, 229–234. [Google Scholar] [CrossRef] [PubMed]
- Ducibella, T.; Schultz, R.M.; Ozil, J.P. Role of calcium signals in early development. Semin. Cell Dev. Biol. 2006, 17, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Colomer, J.; Means, A.R. Physiological roles of the Ca2+/CaM-dependent protein kinase cascade in health and disease. In Sub-Cellular Biochemistry; Springer: Dordrecht, The Netherlands, 2007; Volume 45, pp. 169–214. [Google Scholar]
- Rusciano, M.R.; Salzano, M.; Monaco, S.; Sapio, M.R.; Illario, M.; De Falco, V.; Santoro, M.; Campiglia, P.; Pastore, L.; Fenzi, G.; et al. The Ca2+-calmodulin-dependent kinase II is activated in papillary thyroid carcinoma (PTC) and mediates cell proliferation stimulated by RET/PTC. Endocr. Relat. Cancer 2010, 17, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, N.; Liu, X.; Zheng, Y.; Cao, X. A novel endogenous human CaMKII inhibitory protein suppresses tumor growth by inducing cell cycle arrest via p27 stabilization. J. Biol. Chem. 2008, 283, 11565–11574. [Google Scholar] [CrossRef] [PubMed]
- Yamada, C.; Ozaki, T.; Ando, K.; Suenaga, Y.; Inoue, K.; Ito, Y.; Okoshi, R.; Kageyama, H.; Kimura, H.; Miyazaki, M.; et al. RUNX3 modulates DNA damage-mediated phosphorylation of tumor suppressor p53 at Ser-15 and acts as a co-activator for p53. J. Biol. Chem. 2010, 285, 16693–16703. [Google Scholar] [CrossRef] [PubMed]
- Nevadunsky, N.S.; Barbieri, J.S.; Kwong, J.; Merritt, M.A.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C. RUNX3 protein is overexpressed in human epithelial ovarian cancer. Gynecol. Oncol. 2009, 112, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Stewart, M.; MacKay, N.; Cameron, E.R.; Neil, J.C. The common retroviral insertion locus Dsi1 maps 30 kilobases upstream of the P1 promoter of the murine Runx3/Cbfa3/Aml2 gene. J. Virol. 2002, 76, 4364–4369. [Google Scholar] [CrossRef] [PubMed]
- Young, N.A.; Bruss, M.S.; Gardner, M.; Willis, W.L.; Mo, X.; Valiente, G.R.; Cao, Y.; Liu, Z.; Jarjour, W.N.; Wu, L.C. Oral administration of nano-emulsion curcumin in mice suppresses inflammatory-induced NFkappaB signaling and macrophage migration. PLoS ONE 2014, 9, e111559. [Google Scholar] [CrossRef] [PubMed]
- Whittle, M.C.; Izeradjene, K.; Rani, P.G.; Feng, L.; Carlson, M.A.; DelGiorno, K.E.; Wood, L.D.; Goggins, M.; Hruban, R.H.; Chang, A.E.; et al. RUNX3 Controls a Metastatic Switch in Pancreatic Ductal Adenocarcinoma. Cell 2015, 161, 1345–1360. [Google Scholar] [CrossRef] [PubMed]
- Mariya, T.; Hirohashi, Y.; Torigoe, T.; Asano, T.; Kuroda, T.; Yasuda, K.; Mizuuchi, M.; Sonoda, T.; Saito, T.; Sato, N. Prognostic impact of human leukocyte antigen class I expression and association of platinum resistance with immunologic profiles in epithelial ovarian cancer. Cancer Immunol. Res. 2014, 2, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef] [PubMed]
- American National Standards Institute ANSI/ATCC. Authentication of Human Cell Lines: Standardization of STR Profiling; ASN-0002-2011; ANSI Standards Store: Washington, DC, USA, 2012. [Google Scholar]
- Beaufort, C.M.; Helmijr, J.C.; Piskorz, A.M.; Hoogstraat, M.; Ruigrok-Ritstier, K.; Besselink, N.; Murtaza, M.; van IJcken, W.F.; Heine, A.A.; Smid, M.; et al. Ovarian cancer cell line panel (OCCP): Clinical importance of in vitro morphological subtypes. PLoS ONE 2014, 9, e103988. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Primer | Sequence | Product Size & Annealing Temperature |
|---|---|---|
| CaMKIINα-F | 5′-GACACCAACAACTTCTTCGG-3′ | 84 bp, 58 °C |
| CaMKIINα-R | 5′-CAATAACAACCCGCTTGCT-3′ | |
| HPRT-F | 5′-ACGAAGTGTTGGATATAAGC-3′ | 214 bp, 56 °C |
| HPRT-R | 5′-ATAATTTTACTGGCGATGTC-3′ | |
| ACTB-F | 5′-GGACTTCGAGCAAGAGATGG-3′ | 294 bp, 56 °C |
| ACTB-R | 5′-GCAGTGATCTCCTTCTGCATC-3′ | |
| RUNX3-TV1-F | 5′-ATGGCATCGAACAGCATCTTC-3′ | 337 bp, 60 °C |
| RUNX3-TV1-R | 5′-CCAATGCCACCACCTTGAA-3′ | |
| RUNX3-TV2-F | 5′-ATGCGTATTCCCGTAGACCC-3′ | 295 bp, 60 °C |
| RUNX3-TV2-R | 5′-CCAATGCCACCACCTTGAA-3′ |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heinze, K.; Kritsch, D.; Mosig, A.S.; Dürst, M.; Häfner, N.; Runnebaum, I.B. Functional Analyses of RUNX3 and CaMKIINα in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINα and Dichotomous Roles for RUNX3 Transcript Variants. Int. J. Mol. Sci. 2018, 19, 253. https://doi.org/10.3390/ijms19010253
Heinze K, Kritsch D, Mosig AS, Dürst M, Häfner N, Runnebaum IB. Functional Analyses of RUNX3 and CaMKIINα in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINα and Dichotomous Roles for RUNX3 Transcript Variants. International Journal of Molecular Sciences. 2018; 19(1):253. https://doi.org/10.3390/ijms19010253
Chicago/Turabian StyleHeinze, Karolin, Daniel Kritsch, Alexander S. Mosig, Matthias Dürst, Norman Häfner, and Ingo B. Runnebaum. 2018. "Functional Analyses of RUNX3 and CaMKIINα in Ovarian Cancer Cell Lines Reveal Tumor-Suppressive Functions for CaMKIINα and Dichotomous Roles for RUNX3 Transcript Variants" International Journal of Molecular Sciences 19, no. 1: 253. https://doi.org/10.3390/ijms19010253