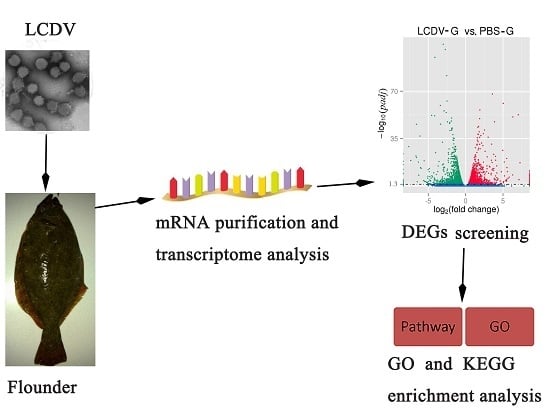
Transcriptome Analysis of Flounder (Paralichthys olivaceus) Gill in Response to Lymphocystis Disease Virus (LCDV) Infection: Novel Insights into Fish Defense Mechanisms
Abstract
:1. Introduction
2. Results
2.1. RNA Quality Verification
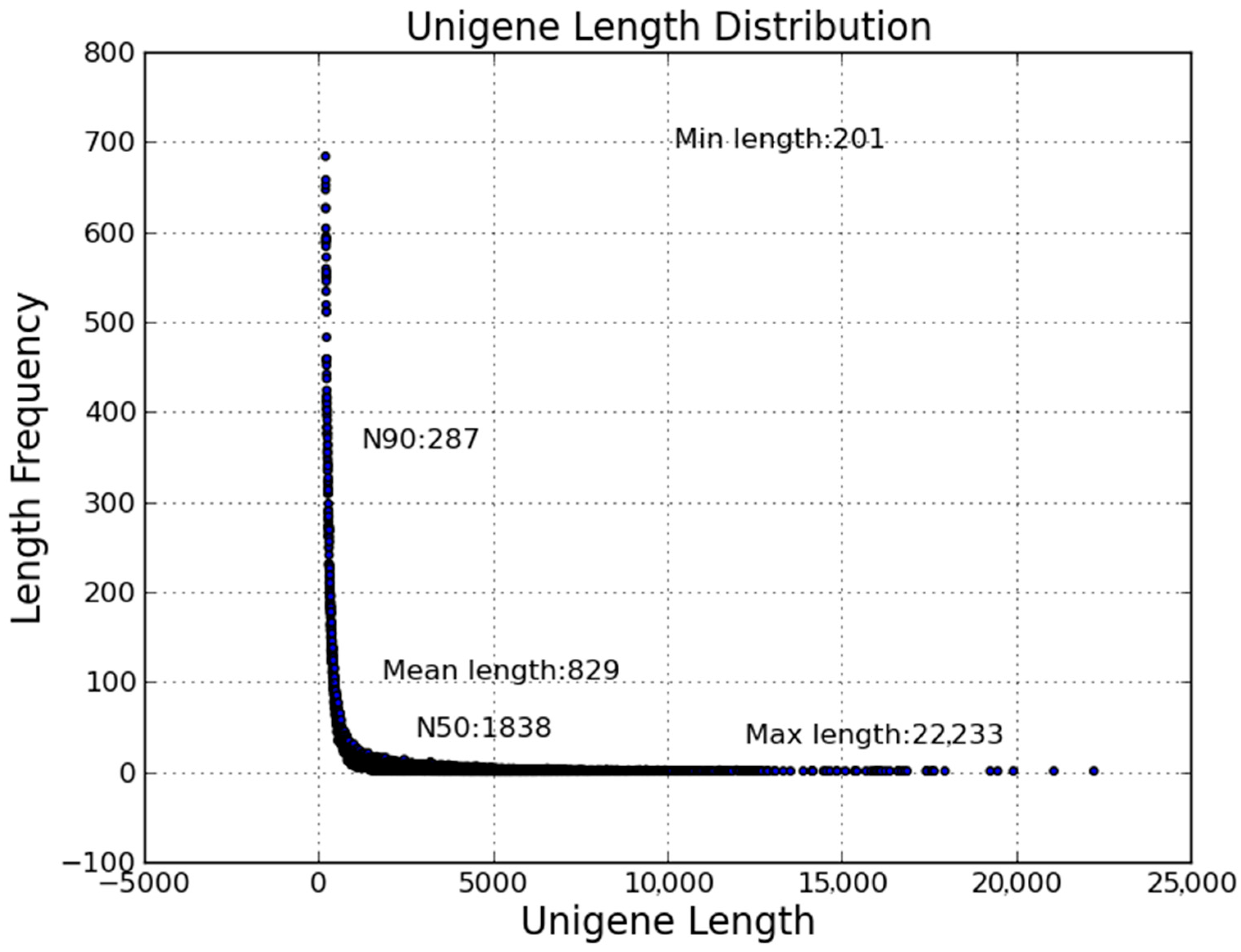
2.2. Transcriptome Sequencing and Assembly
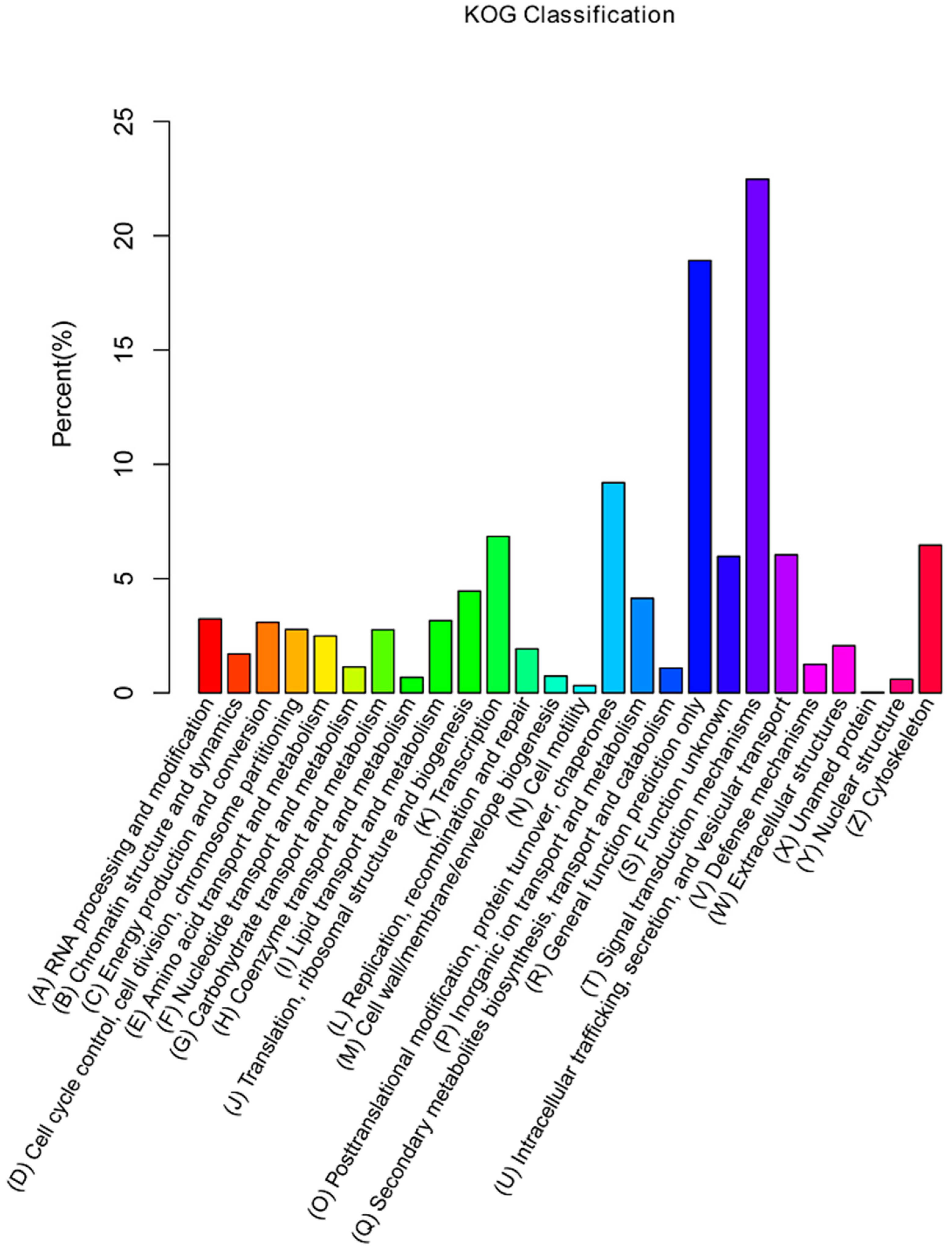
2.3. Functional Annotation of Unigenes Based on GO, KOG, and KEGG Analysis
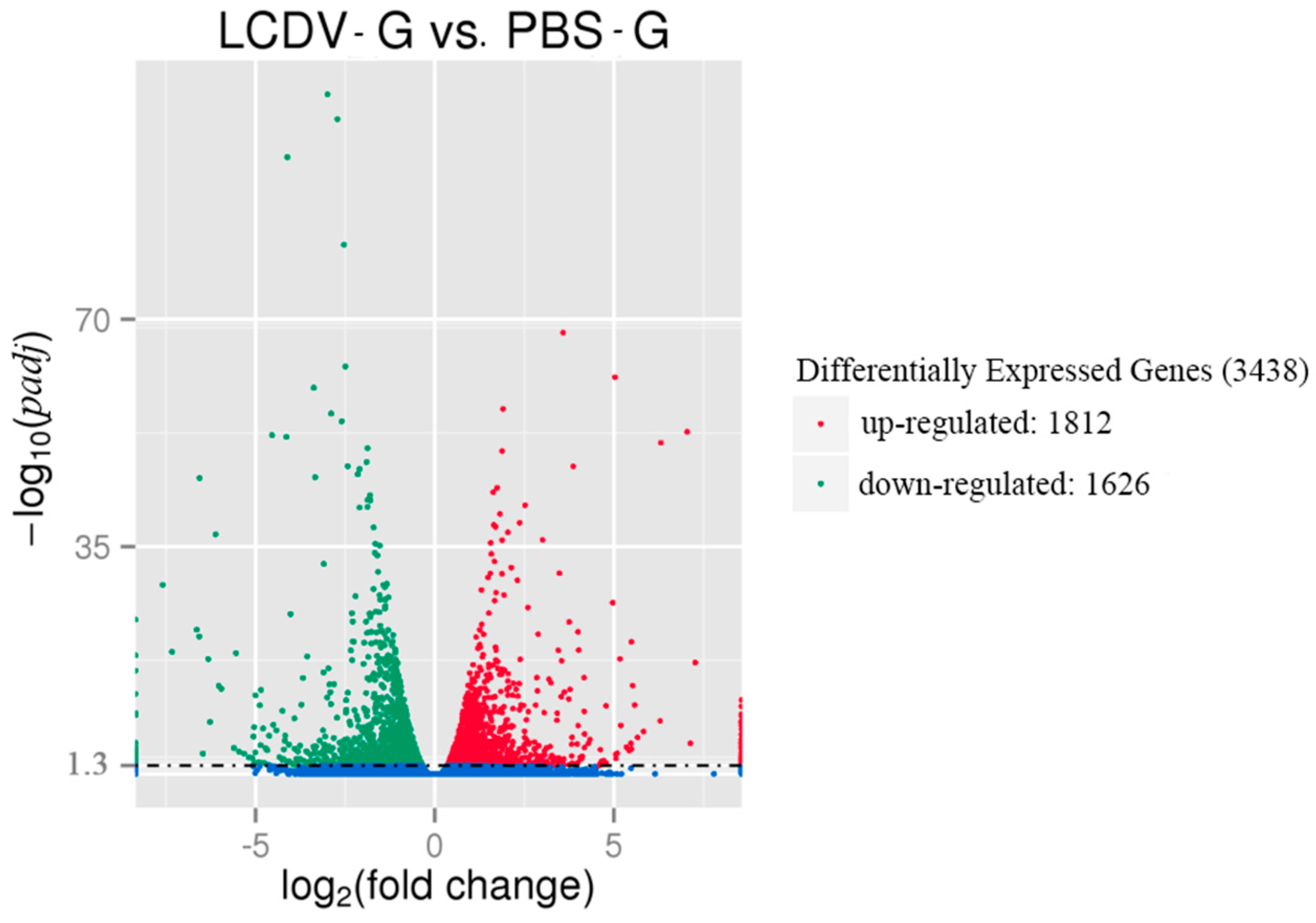
2.4. Differential Gene Expression Induced by LCDV Infection
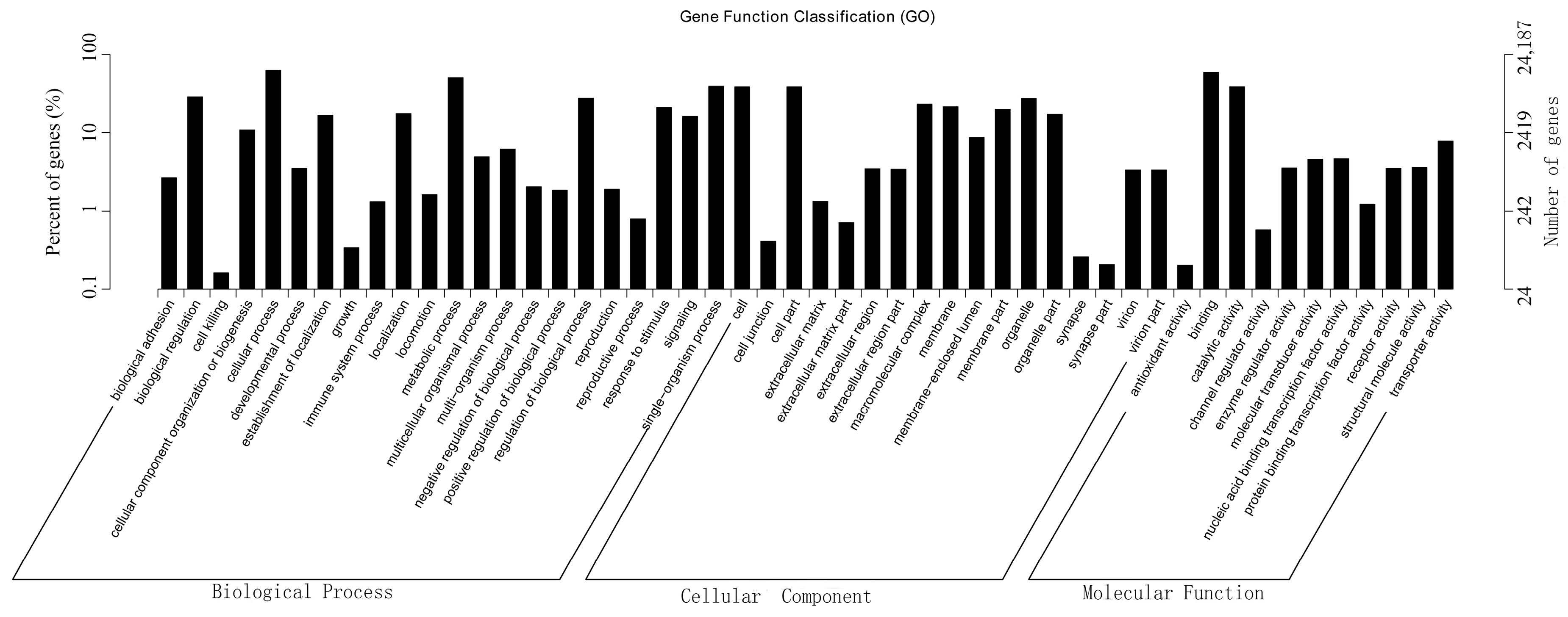
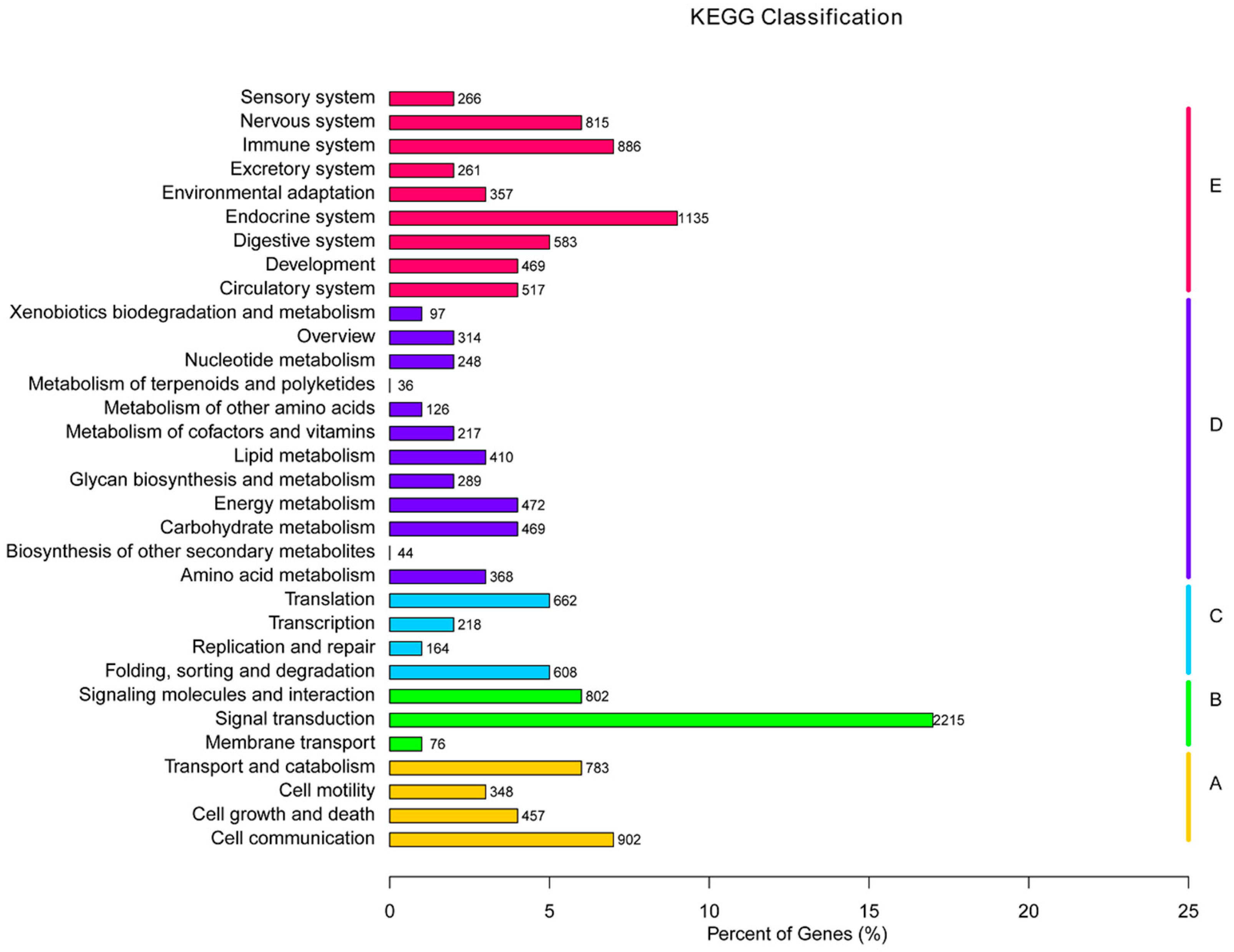
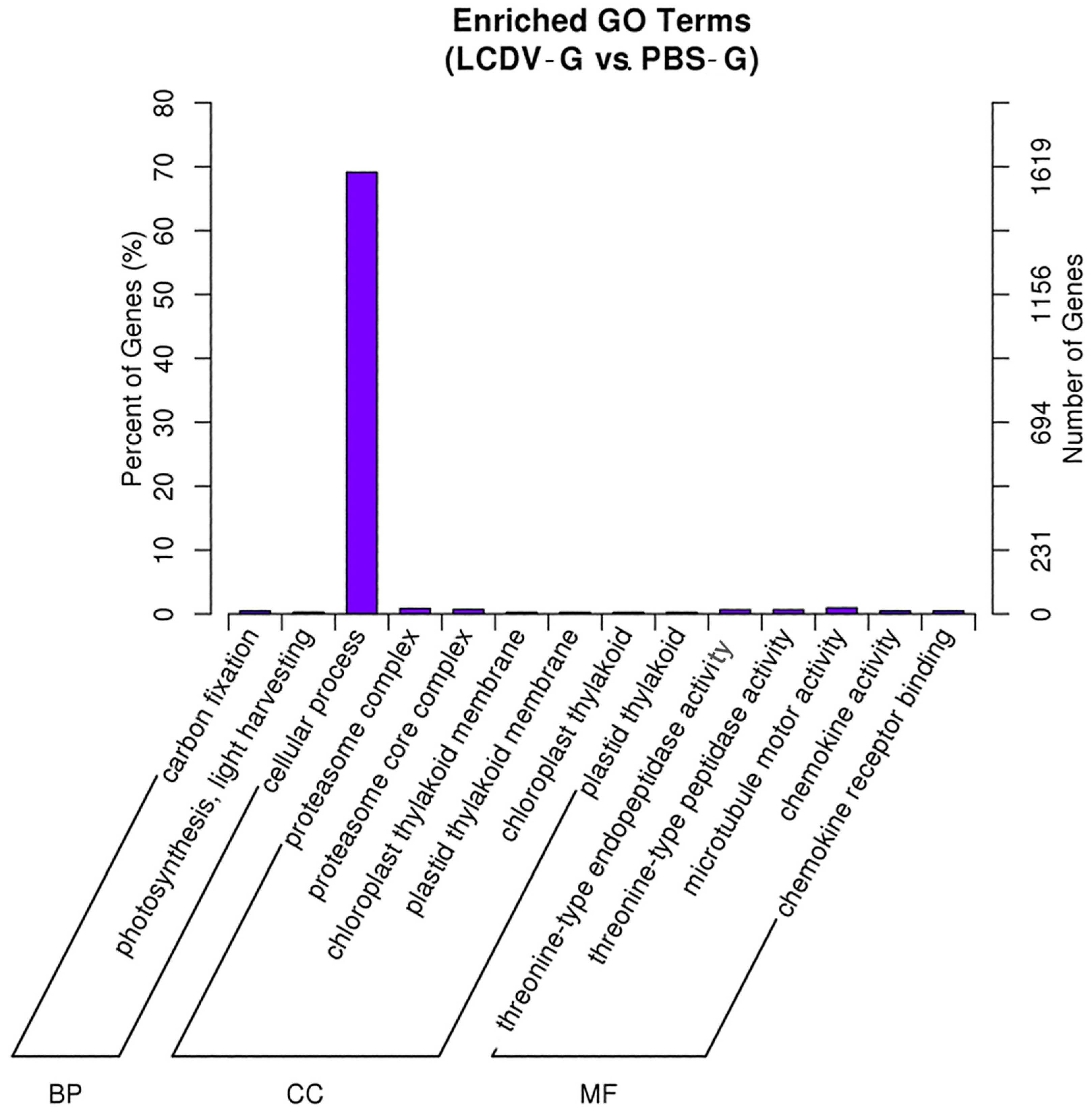
2.5. GO Classification of Differentially Expressed Genes (DEGs)
2.6. Pathway Classification Enrichment of DEGs
2.7. Virus Entry- and Immune-Related DEGs in Response to LCDV Infection
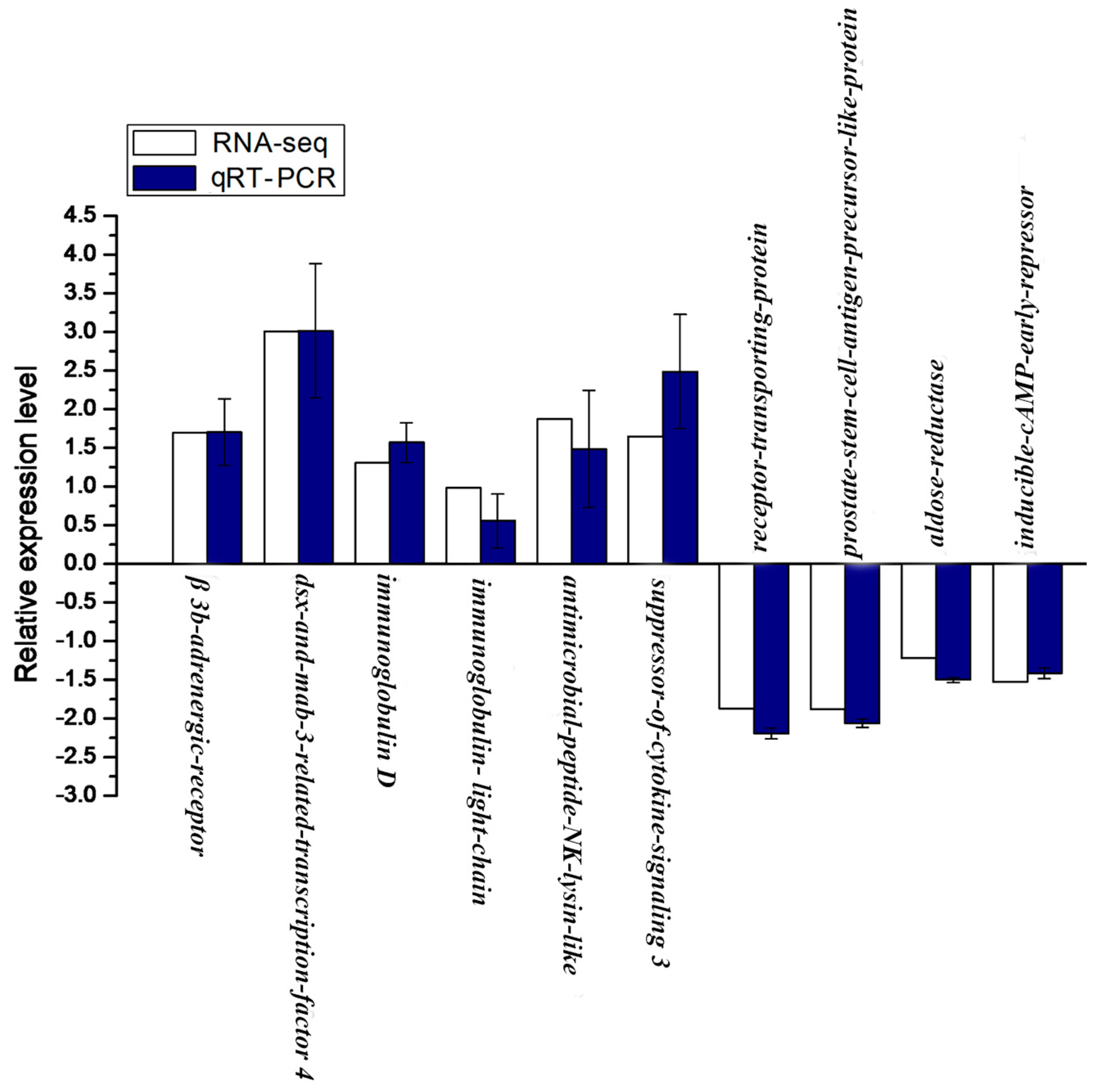
2.8. Validation of RNA-seq Data by qRT-PCR
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Experimental Fish and Virus Infection
4.3. RNA Isolation and Quality Verification
4.4. Library Preparation for Transcriptome Sequencing
4.5. TranScriptome Assembly and Annotation
4.6. Quantification of Gene Expression Levels and Differential Expression Analysis
4.7. GO and KEGG Enrichment Analysis
4.8. Quantitative Real-Time PCR
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kitamura, S.I.; Ko, J.Y.; Lee, W.L.; Kim, S.R.; Song, J.Y.; Kim, D.K.; Oh, M.J. Seasonal prevalence of lymphocystis disease virus and aquabirnavirus in Japanese flounder, Paralichthys olivaceus and blue mussel, Mytilus galloprovincialis. Aquaculture 2007, 266, 26–31. [Google Scholar] [CrossRef]
- Heppell, J.; Berthiaume, L. Ultrastructure of lymphocystis disease virus (LDV) as compared to frog virus 3 (FV3) and chilo iridescent virus (CIV): Effects of enzymatic digestions and detergent degradations. Arch. Virol. 1992, 125, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, R.; Hasegawa, O.; LaPatra, S.; Yoshimizu, M. Isolation and characterization of the Japanese flounder (Paralichthys olivaceus) lymphocystis disease virus. J. Aquat. Anim. Health 2002, 14, 114–123. [Google Scholar] [CrossRef]
- Cheng, S.F.; Zhan, W.B.; Xing, J.; Sheng, X.Z. Development and characterization of monoclonal antibody to the lymphocystis disease virus of Japanese flounder Paralichthys olivaceus isolated from China. J. Virol. Methods 2006, 135, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Pontejo, S.M.; Sánchez, C.; Martín, R.; Mulero, V.; Alcami, A.; Alejo, A. An orphan viral TNF receptor superfamily member identified in lymphocystis disease virus. Virol. J. 2013, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Walker, R. Fine structure of lymphocystis virus of fish. Virology 1962, 18, 503–505. [Google Scholar] [CrossRef]
- Wolf, K. Fish Viruses and Fish Viral Diseases; Cornell University Press: Ithaca, NY, USA, 1988; pp. 268–291. [Google Scholar]
- Zhang, Q.Y.; Xiao, F.; Xie, J.; Li, Z.Q.; Gui, J.F. Complete genome sequence of lymphocystis disease virus isolated from China. J. Virol. 2004, 78, 6982–6994. [Google Scholar] [CrossRef] [PubMed]
- Bowden, R.A.; Oestmann, D.J.; Lewis, D.H.; Frey, M.S. Lymphocystis in red drum. J. Aquat. Anim. Health 1995, 7, 231–235. [Google Scholar] [CrossRef]
- Sheng, X.Z.; Zhan, W.B. Occurrence, development and histochemical characteristics of lymphocystis in cultured Japanese flounder (Paralichthys olivaceus). High Technol. Lett. 2004, 70, 92–96. [Google Scholar]
- Li, Q.; Yue, Z.Q.; Liu, H.; Liang, C.Z.; Zheng, X.L.; Zhao, Y.R. Development and evaluation of a loop-mediated isothermal amplification assay for rapid detection of lymphocystis disease virus. J. Virol. Methods 2010, 163, 378–384. [Google Scholar] [CrossRef] [PubMed]
- Yoshimizu, M.; Iwamoto, R. The Development of Protective Vaccine against Japanese Flounder Lymphocystis Disease Using an Inactivated Virus; Fish Disease Center News, Annual Reports of Fish Disease Control FIDIC: Tokyo, Japan, 2001; pp. 1–14. [Google Scholar]
- Harikrishnan, R.; Balasundaram, C.; Heo, M.S. Effect of probiotics enriched diet on Paralichthys olivaceus infected with lymphocystis disease virus (LCDV). Fish Shellfish Immunol. 2010, 29, 868–874. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Hu, L.L.; Liu, S.S.; Hu, G.B.; Zhang, S.C. Antiviral activity of phosvitin from zebrafish Danio rerio. Dev. Comp. Immunol. 2013, 40, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, S.; Song, J.Y.; Nakayama, K.; Oh, M.J.; Ishida, M.; Kitamura, S.I. Host responses of Japanese flounder Paralichthys olivaceus with lymphocystis cell formation. Fish Shellfish Immunol. 2014, 38, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kozarewa, I.; Ning, Z.; Quail, M.A.; Sanders, M.J.; Berriman, M.; Turner, D.J. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G + C)-biased genomes. Nat. Methods 2009, 6, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Yassour, M.; Kaplan, T.; Fraser, H.B.; Levin, J.Z.; Pfiffner, J.; Adiconis, X.; Schroth, G.; Luo, S.J.; Khrebtukova, I.; Gnirke, A.; et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc. Natl. Acad. Sci. USA 2009, 106, 3264–3269. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, H.J.; Zhao, B.; Sun, Q.Q.; Cao, Y.Y.; Li, R.; Wu, X.X.; Weeda, S.; Li, L.; Ren, S.; et al. The RNA-seq approach to discriminate gene expression profiles in response to melatonin on cucumber lateral root formation. J. Pineal Res. 2014, 56, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Croucher, N.J.; Thomson, N.R. Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 2010, 13, 619–624. [Google Scholar] [CrossRef] [PubMed]
- Marioni, J.C.; Mason, C.E.; Mane, S.M.; Stephens, M.; Gilad, Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008, 18, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; You, F.; Wang, Q.; Weng, S.; Liu, H.; Wang, L.; Zhang, P.; Tan, X. Transcriptional responses of olive flounder (Paralichthys olivaceus) to low temperature. PLoS ONE 2014, 9, e108582. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.H.; Tang, X.Q.; Sheng, X.Z.; Zhan, W.B. Tissue distribution of the 27.8 kDa receptor and its dynamic expression in response to lymphocystis disease virus infection in flounder (Paralichthys olivaceus). J. Comp. Pathol. 2015, 153, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Davidson, G.A.; Lin, S.H.; Secombes, C.J.; Ellis, A.E. Detection of specific and ‘constitutive’ antibody secreting cells in the gills, head kidney and peripheral blood leucocytes of dab (Limanda limanda). Vet. Immunol. Immunopathol. 1997, 58, 363–374. [Google Scholar] [CrossRef]
- Lin, S.H.; Davidson, G.A.; Secombes, C.J.; Ellis, A.E. A morphological study of cells isolated from the perfused gill of dab and Atlantic salmon. J. Fish Biol. 1998, 53, 560–568. [Google Scholar] [CrossRef]
- Cole, A.M.; Darouiche, R.O.; Legarda, D.; Connell, N.; Diamond, G. Characterization of a fish antimicrobial peptide: Gene expression, subcellular localization, and spectrum of activity. Antimicrob. Agents Chemother. 2000, 44, 2039–2045. [Google Scholar] [CrossRef] [PubMed]
- Merino-Contreras, M.L.; Guzman-Murillo, M.A.; Ruiz-Bustos, E.; Romero, M.J.; Cadena-Roa, M.A.; Ascencio, F. Mucosal immune response of spotted sand bass Paralabrax maculatofasciatus (Steindachner, 1868) orally immunised with an extracellular lectin of Aeromonas veronii. Fish Shellfish Immunol. 2001, 11, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Romøren, K.; Fjeld, X.T.; Poléo, A.; Smistad, G.; Thu, B.J.; Evensen, Ø. Transfection efficiency and cytotoxicity of cationic liposomes in primary cultures of rainbow trout (Oncorhynchus mykiss) gill cells. Biochim. Biophys. Acta Biomembr. 2005, 1717, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Gomez, D.; Sunyer, J.O.; Salinas, I. The mucosal immune system of fish: The evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35, 1729–1739. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Jeong, B.M.; Kim, W.S.; Kim, J.O.; Lim, C.H.; Cho, J.Y.; Eun, J.B.; Moon, J.H.; Kim, S.R.; Park, M.A.; et al. Detection of lymphocystis disease virus (LCDV) in olive flounder (Paralichthys olivaceus) using efficient extraction and concentration methods. Food Sci. Biotechnol. 2010, 19, 1693–1696. [Google Scholar] [CrossRef]
- Zhan, W.B.; Li, Y.Q.; Sheng, X.Z.; Xing, J.; Tang, X.Q. Detection of lymphocystis disease virus in Japanese flounder Paralichthys olivaceus and other marine teleosts from northern China. Chin. J. Oceanol. Limnol. 2010, 28, 1213–1220. [Google Scholar] [CrossRef]
- Sheng, X.Z.; Wang, M.; Xing, J.; Zhan, W.B. Monoclonal antibodies against 27.8 kDa protein receptor efficiently block lymphocystis disease virus infection in flounder Paralichthys olivaceus gill cells. Dis. Aquat. Org. 2012, 100, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.Q.; Diao, J.; Zhan, W.B.; Sheng, X.Z. Identification of a 37.6 kDa membrane protein from flounder gill cells involved in binding and infection to lymphocystis disease virus. Aquaculture 2012, 358, 132–138. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Lyles, D.S. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 2000, 64, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Chiou, P.P.; Kim, C.H.; Ormonde, P.; Leong, J.A.C. Infectious hematopoietic necrosis virus matrix protein inhibits host-directed gene expression and induces morphological changes of apoptosis in cell cultures. J. Virol. 2000, 74, 7619–7627. [Google Scholar] [CrossRef] [PubMed]
- Purcell, M.K.; Marjara, I.S.; Batts, W.; Kurath, G.; Hansen, J.D. Transcriptome analysis of rainbow trout infected with high and low virulence strains of infectious hematopoietic necrosis virus. Fish Shellfish Immunol. 2011, 30, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Gale, M.; Tan, S.L.; Katze, M.G. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 2000, 64, 239–280. [Google Scholar] [CrossRef] [PubMed]
- Charo, I.F. Ransohoff, R.M. The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 2006, 354, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Livnat-Levanon, N.; Glickman, M.H. Ubiquitin–proteasome system and mitochondria-reciprocity. Biochim. Biophys. Acta Gene Regul. Mech. 2011, 1809, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Orlowski, R.Z. The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death Differ. 1999, 6, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sheng, X.Z.; Xing, J.; Tang, X.Q.; Zhan, W.B. Identification of a 27.8 kDa protein from flounder gill cells involved in lymphocystis disease virus binding and infection. Dis. Aquat. Org. 2011, 94, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.B.; Cong, R.S.; Fan, T.J.; Mei, X.G. Induction of apoptosis in a flounder gill cell line by lymphocystis disease virus infection. J. Fish Dis. 2004, 27, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhou, W.W.; Zhou, Y.J.; Wu, J.X.; Zhou, X.P. Transcriptome and comparative gene expression analysis of Sogatella furcifera (Horváth) in response to Southern rice black-streaked dwarf virus. PLoS ONE 2012, 7, e36238. [Google Scholar] [CrossRef] [PubMed]
- Sayers, T.J. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol. Immunother. 2011, 60, 1173–1180. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Kawai, Y.; Fergusson, M.M.; Rovira, I.I.; Bishop, A.J.; Motoyama, N.; Cao, L.; Finkel, T. Atg7 modulates p53 activity to regulate cell cycle and survival during metabolic stress. Science 2012, 336, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zhang, Z.S.; Zhang, Y.L.; Zhou, D.Y. Curcumin inhibits cell proliferation by interfering with the cell cycle and inducing apoptosis in colon carcinoma cells. Anticancer Res. 1998, 19, 3675–3680. [Google Scholar]
- Castellano, M.M.; Boniotti, M.B.; Caro, E.; Schnittger, A.; Gutierrez, C. DNA replication licensing affects cell proliferation or endoreplication in a cell type—Specific manner. Plant Cell 2004, 16, 2380–2393. [Google Scholar] [CrossRef] [PubMed]
- Menon, V.; Povirk, L. Involvement of p53 in the repair of DNA double strand breaks: Multifaceted roles of p53 in homologous recombination repair (HRR) and non-homologous end joining (NHEJ). In Mutant p53 and MDM2 in Cancer; Springer: Dordrecht, The Netherlands, 2014; pp. 321–336. [Google Scholar]
- Hollstein, M.; Sidransky, D.; Vogelstein, B.; Harris, C.C. p53 mutations in human cancers. Science 1991, 253, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Van Kooyk, Y.; Rabinovich, G.A. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat. Immunol. 2008, 9, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Aamelfot, M.; Dale, O.B.; Weli, S.C.; Koppang, E.O.; Falk, K. Expression of the infectious salmon anemia virus receptor on Atlantic salmon endothelial cells correlates with the cell tropism of the virus. J. Virol. 2012, 86, 10571–10578. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.Z.; Wu, R.H.; Tang, X.Q.; Xing, J.; Zhan, W.B. Tissue Localization of Lymphocystis Disease Virus (LCDV) Receptor-27.8 kDa and Its Expression Kinetics Induced by the Viral Infection in Turbot (Scophthalmus maximus). Int. J. Mol. Sci. 2015, 16, 26506–26519. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Kaisho, T.; Akira, S. Toll-like receptors. Annu. Rev. Immunol. 2003, 21, 335–376. [Google Scholar] [CrossRef] [PubMed]
- Schnare, M.; Barton, G.M.; Holt, A.C.; Takeda, K.; Akira, S.; Medzhitov, R. Toll-like receptors control activation of adaptive immune responses. Nat. Immunol. 2001, 2, 947–950. [Google Scholar] [CrossRef] [PubMed]
- Pasare, C.; Medzhitov, R. Toll pathway-dependent blockade of CD4+ CD25+ T cell-mediated suppression by dendritic cells. Science 2003, 299, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Damania, B. Upregulation of the TLR3 pathway by Kaposi’s sarcoma-associated herpesvirus during primary infection. J. Virol. 2008, 82, 5440–5449. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.D.; Kondo, H.; Hirono, I.; Aoki, T. Molecular cloning and characterization of Toll-like receptor 14 in Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2011, 30, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Proinflammatory cytokines. Chest J. 2000, 118, 503–508. [Google Scholar] [CrossRef]
- Görgen, I.; Hartung, T.H.O.M.A.S.; Leist, M.A.R.C.E.L.; Niehörster, M.; Tiegs, G.I.S.A.; Uhlig, S.; Weitzel, F.; Wendel, A. Granulocyte colony-stimulating factor treatment protects rodents against lipopolysaccharide-induced toxicity via suppression of systemic tumor necrosis factor-alpha. J. Immunol. 1992, 149, 918–924. [Google Scholar] [PubMed]
- Boneberg, E.M.; Hartung, T. Molecular aspects of anti-inflammatory action of G-CSF. Inflamm. Res. 2002, 51, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Sutton, V.R.; Vaux, D.L.; Trapani, J.A. Bcl-2 prevents apoptosis induced by perforin and granzyme B, but not that mediated by whole cytotoxic lymphocytes. J. Immunol. 1997, 158, 5783–5790. [Google Scholar] [PubMed]
- Becker, T.C.; Wherry, E.J.; Boone, D.; Murali-Krishna, K.; Antia, R.; Ma, A.; Ahmed, R. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 2002, 195, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.A.; Hermogenes, A.W.; Sacks, H.S.; Lau, J.; Levandowski, R.A. The efficacy of influenza vaccine in elderly persons. A meta-analysis and review of the literature. Ann. Int. Med. 1995, 123, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, N.; Seder, R.A.; Reddy, A.; Feldman, M.V. IL-12 in conjunction with dendritic cells enhances antiviral CD8+ CTL responses in vitro. J. Clin. Investig. 1996, 98, 715. [Google Scholar] [CrossRef] [PubMed]
- Netea, M.G.; van der Graaf, C.; van der Meer, J.W.; Kullberg, B.J. Toll-like receptors and the host defense against microbial pathogens: Bringing specificity to the innate-immune system. J. Leukoc. Biol. 2004, 75, 749–755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Sun, B.; Luo, Y.; Zhang, Y.; Nie, P. Distribution of IgM, IgD and IgZ in mandarin fish, Siniperca chuatsi lymphoid tissues and their transcriptional changes after Flavobacterium columnare stimulation. Aquaculture 2009, 288, 14–21. [Google Scholar] [CrossRef]
- Edholm, E.S.; Bengten, E.; Stafford, J.L.; Sahoo, M.; Taylor, E.B.; Miller, N.W. Identification of two IgD+ B cell populations in channel catfish, Ictalurus punctatus. J. Immunol. 2010, 185, 4082–4094. [Google Scholar] [CrossRef] [PubMed]
- Castro, R.; Jouneau, L.; Pham, H.P.; Bouchez, O.; Giudicellio, V.; Lefranc, M.P.; Quillet, E.A.; Benmansour, F.; Cazals, A.; Six, S.; et al. Teleost Fish mount complex clonal IgM and IgT responses in spleen upon systemic viral infection. PLoS Pathog. 2013, 9, e1003098. [Google Scholar] [CrossRef] [PubMed]
- Mazanec, M.B.; Nedrud, J.G.; Kaetzel, C.S.; Lamm, M.E. A three-tiered view of the role of IgA in mucosal defense. Immunol. Today 1993, 14, 430–435. [Google Scholar] [CrossRef]
- Lamm, M.E. Interaction of antigens and antibodies at mucosal surfaces. Annu. Rev. Microbiol. 1997, 51, 311–340. [Google Scholar] [CrossRef] [PubMed]
- Kaetzel, C.S. Polymeric Ig receptor: Defender of the fort or Trojan horse? Curr. Microbiol. 2001, 11, 35–38. [Google Scholar] [CrossRef]
- Kaetzel, C.S. The polymeric immunoglobulin receptor: Bridging innate and adaptive immune responses at mucosal surfaces. Immunol. Rev. 2005, 206, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.J.; Zhan, W.B.; Ding, B.J.; Sheng, X.Z. Molecular cloning and expression analysis of polymeric immunoglobulin receptor in flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2013, 35, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Kortum, A.N.; Rodriguez-Nunez, I.; Yang, J.; Shim, J.; Runft, D.; O’Driscoll, M.L.; Haire, R.N.; Cannon, J.P.; Turner, P.M.; Litman, R.T.; et al. Differential expression and ligand binding indicate alternative functions for zebrafish polymeric immunoglobulin receptor (pIgR) and a family of pIgR-like (PIGRL) proteins. Immunogenetics 2014, 66, 267–279. [Google Scholar] [CrossRef] [PubMed]
- Pal, K.; Kaetzel, C.S.; Brundage, K.; Cunningham, C.A.; Cuff, C.F. Regulation of polymeric immunoglobulin receptor expression by reovirus. J. Gen. Virol. 2005, 86, 2347–2357. [Google Scholar] [CrossRef] [PubMed]
- Byon, J.Y.; Ohira, T.; Hirono, I.; Aoki, T. Comparative immune responses in Japanese flounder, Paralichthys olivaceus after vaccination with viral hemorrhagic septicemia virus (VHSV) recombinant glycoprotein and DNA vaccine using a microarray analysis. Vaccine 2006, 24, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Huang, X.; Yan, Y.; Cai, J.; Ouyang, Z.; Cui, H.; Wang, P.; Qin, Q. Transcriptome analysis of orange-spotted grouper (Epinephelus coioides) spleen in response to Singapore grouper iridovirus. BMC Genom. 2011, 12, 556. [Google Scholar] [CrossRef] [PubMed]
- Broering, R.; Zhang, X.; Kottilil, S.; Trippler, M.; Jiang, M.; Lu, M.; Gerken, G.; Schlaak, J.F. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut 2010, 59, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Avunje, S.; Oh, M.J.; Jung, S.J. Impaired TLR2 and TLR7 response in olive flounder infected with viral haemorrhagic septicaemia virus at host susceptible 15 C but high at non-susceptible 20 C. Fish Shellfish Immunol. 2013, 34, 1236–1243. [Google Scholar] [CrossRef] [PubMed]
- Carballo, C.; Castro, D.; Borrego, J.J.; Manchado, M. Gene expression profiles associated with lymphocystis disease virus (LCDV) in experimentally infected Senegalese sole (Solea senegalensis). Fish Shellfish Immunol. 2017, 66, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Kilkenny, C.; Browne, W.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Animal research: Reporting in vivo experiments: The ARRIVE guidelines. J. Gene Med. 2010, 12, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
| Sample Name | OD260/OD280 | 28S/18S RNA Ratio | RIN Value |
|---|---|---|---|
| PBS-G1 | 1.853 ± 0.086 | 1.7 ± 0.3 | 8.9 ± 0.3 |
| PBS-G2 | 1.887 ± 0.104 | 1.8 ± 0.2 | 8.7 ± 0.2 |
| LCDV-G1 | 1.971 ± 0.018 | 2.1 ± 0.1 | 9.1 ± 0.1 |
| LCDV-G2 | 1.865 ± 0.117 | 1.9 ± 0.2 | 9.3 ± 0.2 |
| Category | PBS-G1 | PBS-G2 | LCDV-G1 | LCDV-G2 | Summary |
|---|---|---|---|---|---|
| Raw reads | 47,382,188 | 41,257,434 | 55,497,824 | 59,422,436 | 203,559,882 |
| Clean reads | 46,280,260 | 40,363,354 | 51,413,710 | 55,167,846 | 193,225,170 |
| Q20 (%) | 93.12–95.21 | 93.13–95.29 | 92.05–96.24 | 91.72–96.35 | |
| GC (%) | 47.69–47.71 | 47.51–47.53 | 47.41–47.55 | 47.61–47.76 | |
| Total mapped | 39,092,630 (84.47%) | 34,395,222 (85.21%) | 43,059,442 (83.75%) | 46,511,408 (84.31%) |
| Annotation in Database | Number of Unigenes | Percentage (%) |
|---|---|---|
| Annotated in Nr | 24,922 | 23.44 |
| Annotated in Nt | 27,528 | 25.89 |
| Annotated in KEGG Ortholog | 12,881 | 12.11 |
| Annotated in Swiss-Prot | 21,572 | 20.29 |
| Annotated in Pfam | 23,245 | 21.86 |
| Annotated in GO | 24,187 | 22.75 |
| Annotated in KOG | 13,871 | 13.04 |
| Annotated in all databases | 8207 | 7.72 |
| Annotated in at least one database | 36,537 | 34.37 |
| Total unigenes | 106,293 | 100 |
| Pathways | Pathways ID | Branch in the KEGG Database | Up-/Down-Regulated Genes |
|---|---|---|---|
| Proteasome | ko03050 | genetic information processing | 25/0 |
| Base excision repair | ko03410 | genetic information processing | 9/1 |
| Homologous recombination | ko03440 | genetic information processing | 7/2 |
| DNA replication | ko03030 | genetic information processing | 12/0 |
| Mismatch repair | ko03430 | genetic information processing | 7/0 |
| Spliceosome | ko03040 | genetic information processing | 22/3 |
| Aminoacyl-tRNA biosynthesis | ko00970 | genetic information processing | 10/0 |
| Nucleotide excision repair | ko03420 | genetic information processing | 10/0 |
| Metabolic pathways | ko01100 | metabolism | 128/77 |
| Glutathione metabolism | ko00480 | metabolism | 11/3 |
| Other types of O-glycan biosynthesis | ko00514 | metabolism | 6/4 |
| Amino sugar and nucleotide sugar metabolism | ko00520 | metabolism | 9/7 |
| p53 signaling pathway | ko04115 | cellular processes | 15/8 |
| Cell cycle | ko04110 | cellular processes | 27/8 |
| Tumor necrosis factor (TNF) signaling pathway | ko04668 | environmental information processing | 16/9 |
| Gene Name | Expression after Infection | Expression in Control | Ratio of Expression (Infection vs. Control) |
|---|---|---|---|
| β actin | 12,056 | 7655 | 1.575 |
| TLR3 | 38 | 4 | 9.500 |
| TLR14 | 409 | 1259 | 0.325 |
| interferon regulatory factor 2 | 286 | 138 | 2.072 |
| interferon regulatory factor | 15,252 | 9252 | 1.649 |
| interferon γ receptor α chain | 5521 | 3731 | 1.480 |
| interferon γ | 145 | 68 | 2.132 |
| interferon regulatory factor 8 | 355 | 223 | 1.592 |
| interleukin-8 receptor | 217 | 49 | 4.429 |
| interleukin-12 subunit β-like | 564 | 315 | 1.790 |
| Interleukin-15 | 413 | 242 | 1.707 |
| chemokine XC receptor 1-like | 589 | 216 | 2.727 |
| CC chemokine | 124 | 24 | 5.167 |
| C-C motif chemokine 3 precursor | 218 | 85 | 2.565 |
| CXC chemokine receptor 4 | 4244 | 2835 | 1.497 |
| CXC chemokine receptor 3 | 1009 | 634 | 1.591 |
| C-C motif chemokine 4 precursor | 284 | 158 | 1.797 |
| CXC chemokine | 1393 | 979 | 1.423 |
| chemokine XC receptor 1-like | 787 | 537 | 1.466 |
| C-X-C chemokine receptor type 4 protein | 166 | 90 | 1.844 |
| C-C motif chemokine 20 precursor | 111 | 361 | 0.307 |
| C-C motif chemokine 4-like | 26 | 115 | 0.226 |
| C-C chemokine receptor type 2-like | 82 | 207 | 0.396 |
| TNF ligand superfamily member 13B | 883 | 405 | 2.180 |
| TNF ligand superfamily member 6-like | 553 | 305 | 1.813 |
| TNF alpha-induced protein 2-like | 4776 | 2516 | 1.898 |
| TNF ligand superfamily member 14-like | 666 | 452 | 1.473 |
| TNF receptor-1 | 1186 | 3661 | 0.324 |
| TNF ligand superfamily member 13B | 883 | 405 | 2.180 |
| TNF ligand superfamily member 6-like | 553 | 305 | 1.813 |
| TNF ligand superfamily member 14-like | 666 | 452 | 1.473 |
| granulocyte colony-stimulating factor | 21 | 129 | 0.163 |
| perforin | 2053 | 1136 | 1.807 |
| natural killer enhancing factor | 1209 | 628 | 1.925 |
| bcl-2 | 402 | 641 | 0.627 |
| granzyme II | 3392 | 1213 | 2.796 |
| IgD | 47,080 | 19,068 | 2.469 |
| pIgR | 202 | 123 | 1.642 |
| ISG 15 | 2199 | 3851 | 0.571 |
| Mx | 932 | 2203 | 0.423 |
| antimicrobial peptide NK-lysin-like | 1361 | 372 | 3.659 |
| complement factor D | 16,310 | 8844 | 1.844 |
| complement component C3 | 123 | 61 | 2.016 |
| complement component C6-like | 197 | 57 | 3.456 |
| ubiquitin-conjugating enzyme E2 A | 352 | 221 | 1.593 |
| ubiquitin-conjugating enzyme E2 C-like | 340 | 142 | 2.394 |
| E3 ubiquitin-protein ligase CHFR-like | 2191 | 1409 | 1.555 |
| E3 ubiquitin-protein ligase RING2-like | 1023 | 701 | 1.459 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, R.; Sheng, X.; Tang, X.; Xing, J.; Zhan, W. Transcriptome Analysis of Flounder (Paralichthys olivaceus) Gill in Response to Lymphocystis Disease Virus (LCDV) Infection: Novel Insights into Fish Defense Mechanisms. Int. J. Mol. Sci. 2018, 19, 160. https://doi.org/10.3390/ijms19010160
Wu R, Sheng X, Tang X, Xing J, Zhan W. Transcriptome Analysis of Flounder (Paralichthys olivaceus) Gill in Response to Lymphocystis Disease Virus (LCDV) Infection: Novel Insights into Fish Defense Mechanisms. International Journal of Molecular Sciences. 2018; 19(1):160. https://doi.org/10.3390/ijms19010160
Chicago/Turabian StyleWu, Ronghua, Xiuzhen Sheng, Xiaoqian Tang, Jing Xing, and Wenbin Zhan. 2018. "Transcriptome Analysis of Flounder (Paralichthys olivaceus) Gill in Response to Lymphocystis Disease Virus (LCDV) Infection: Novel Insights into Fish Defense Mechanisms" International Journal of Molecular Sciences 19, no. 1: 160. https://doi.org/10.3390/ijms19010160