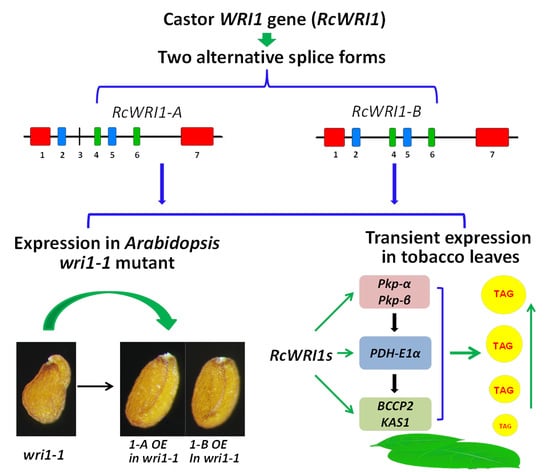
Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves
Abstract
:1. Introduction
2. Results
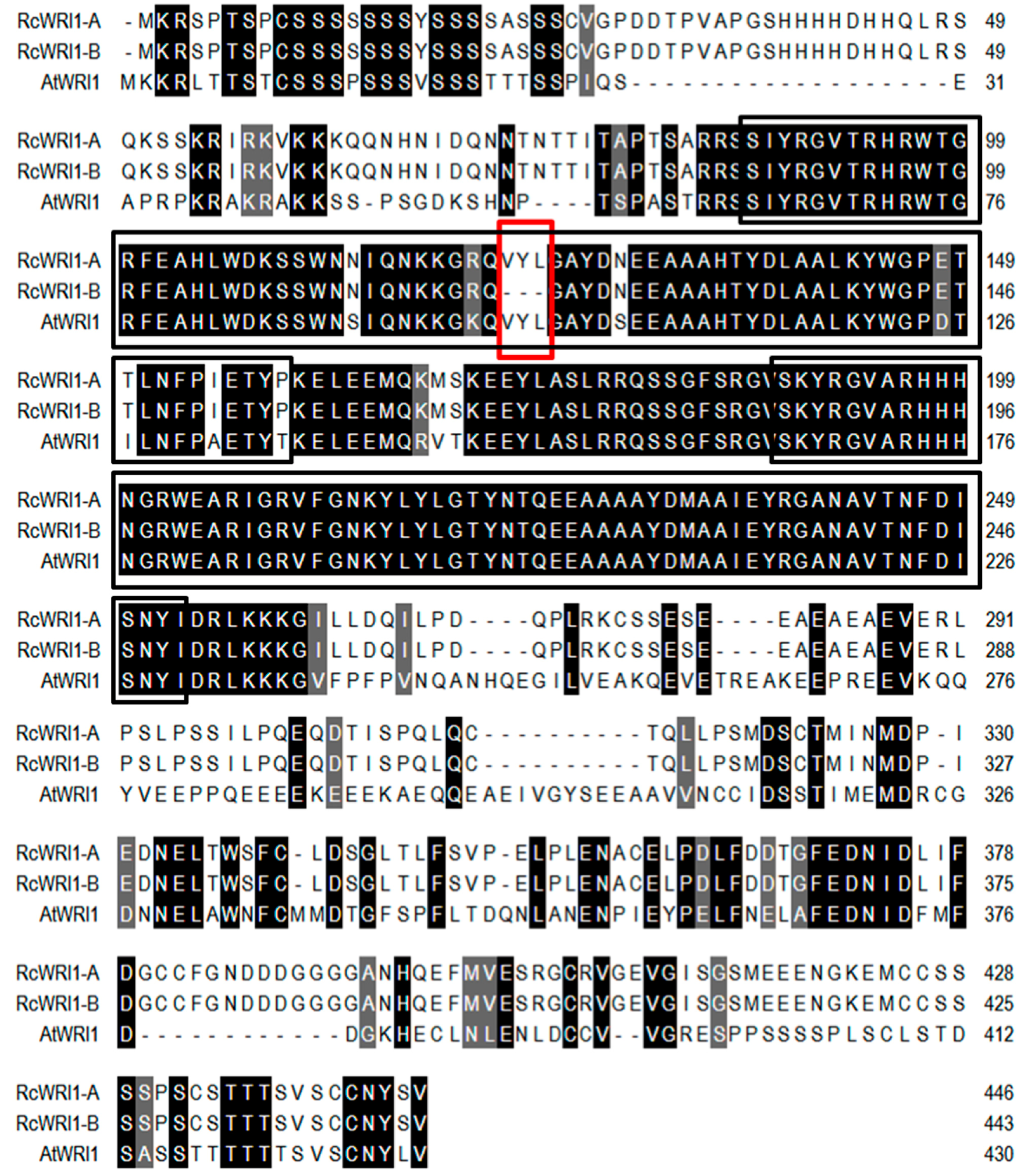
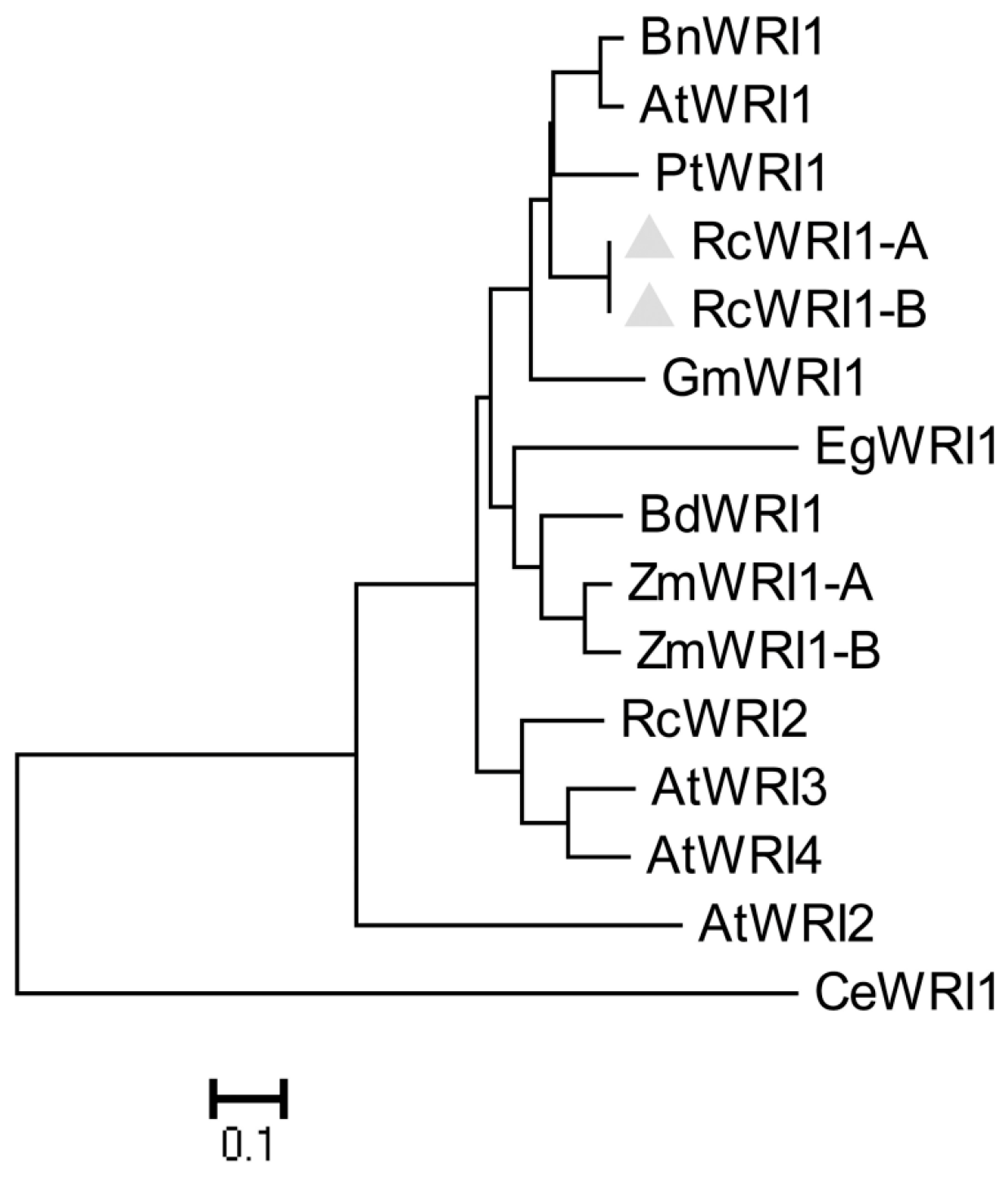
2.1. Molecular Characterization of Castor RcWRI1
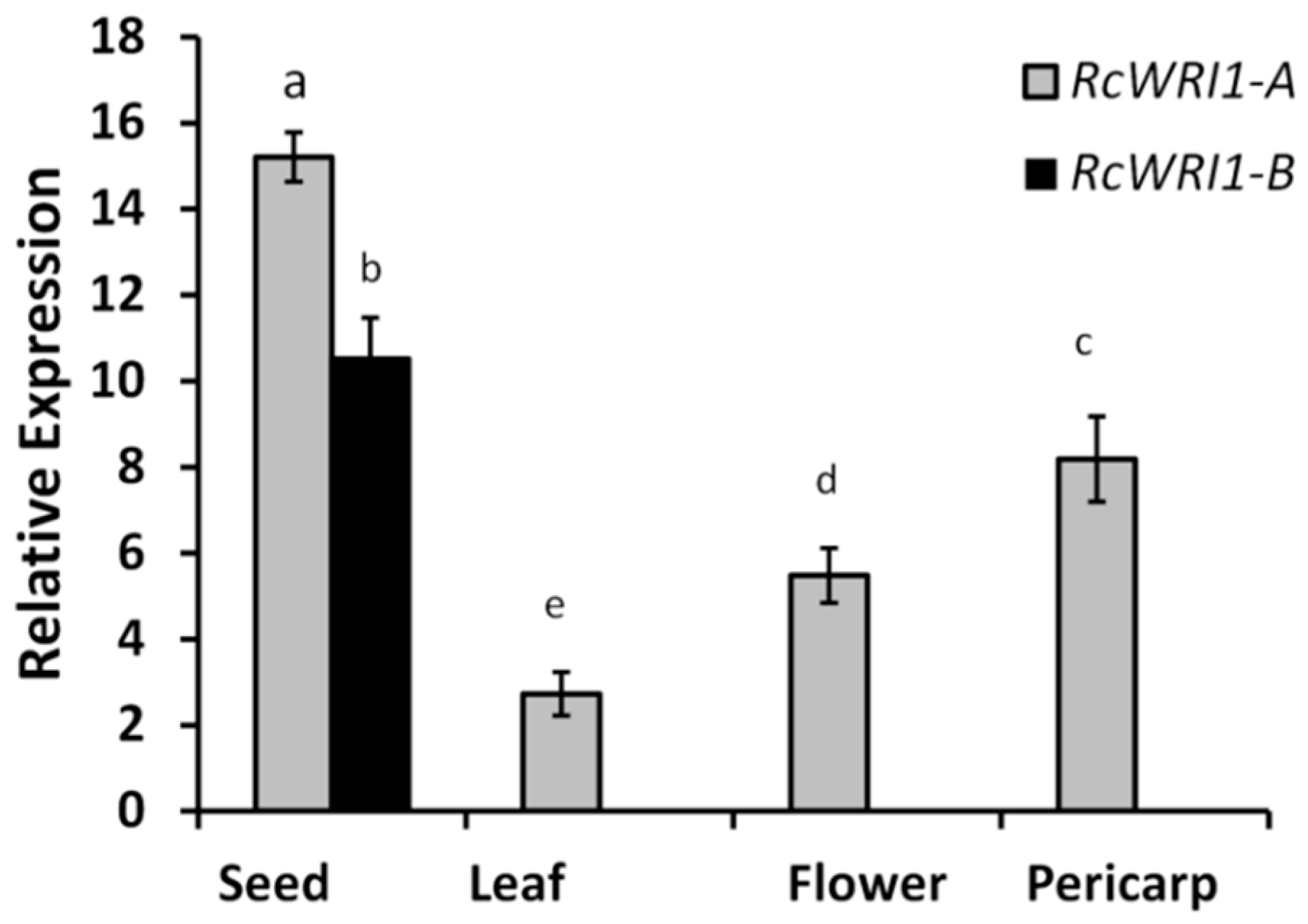
2.2. Expression Profiles of RcWRI1-A and RcWRI1-B in Various Castor Organs
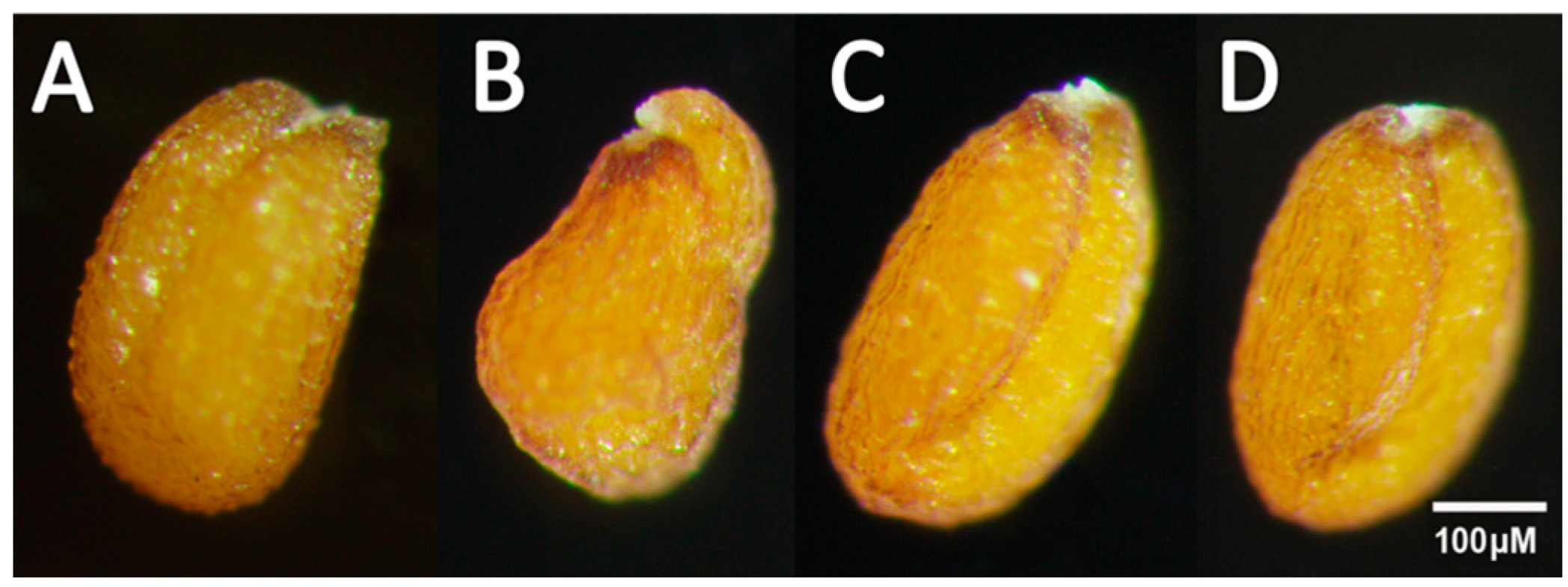
2.3. Overexpression of Individual RcWRI1 Splice Forms Restored Seed Total Lipid Content in the Arabidopsis wri1-1 Mutant
2.4. Transient Expression of RcWRI1s Enhances the Expression of the Downstream Genes in the Fatty Acid Biosynthesis Pathway
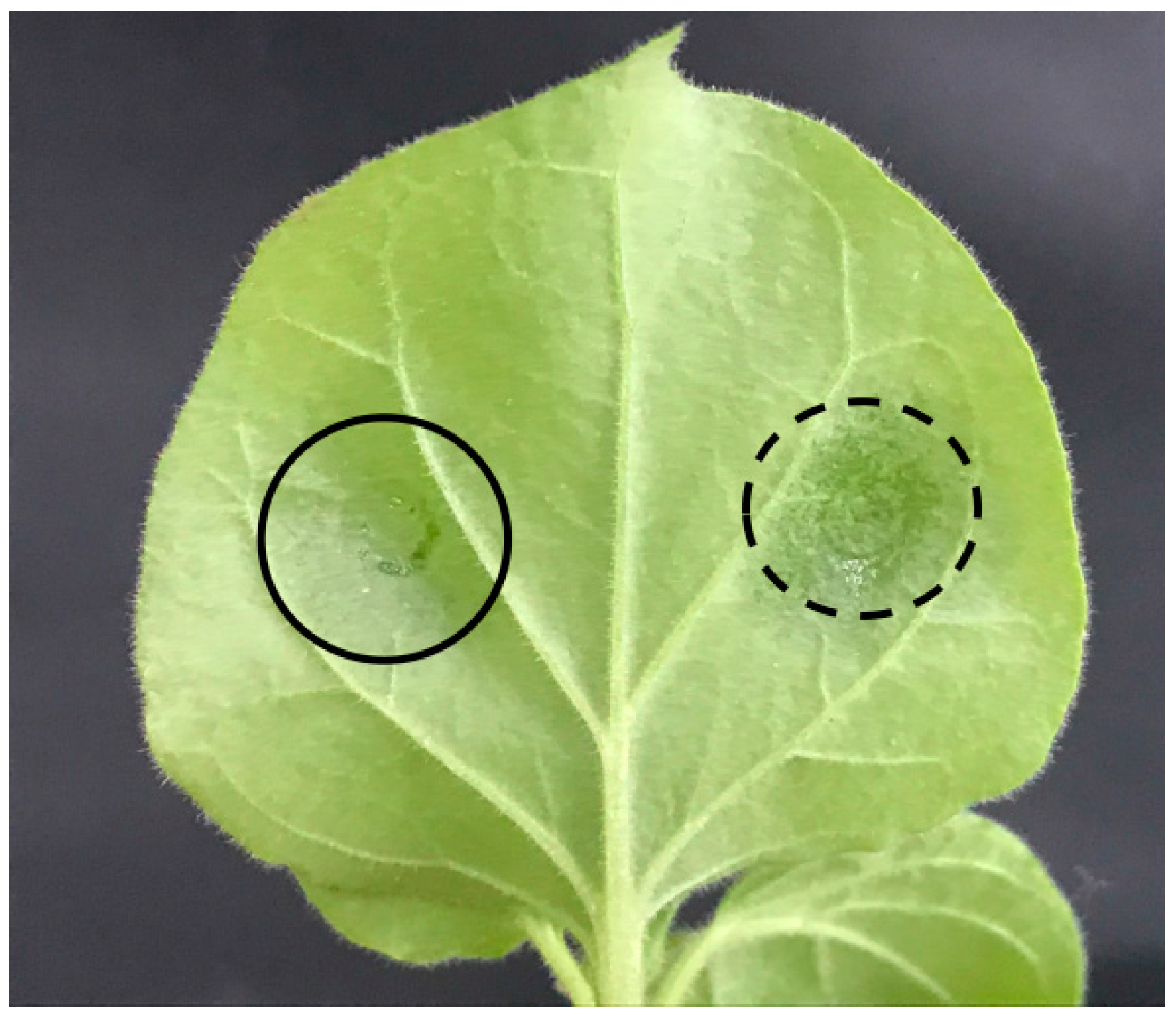
2.5. Ectopic Expression of RcWRI1s Increases Total Lipid Content in N. benthamiana Leaves
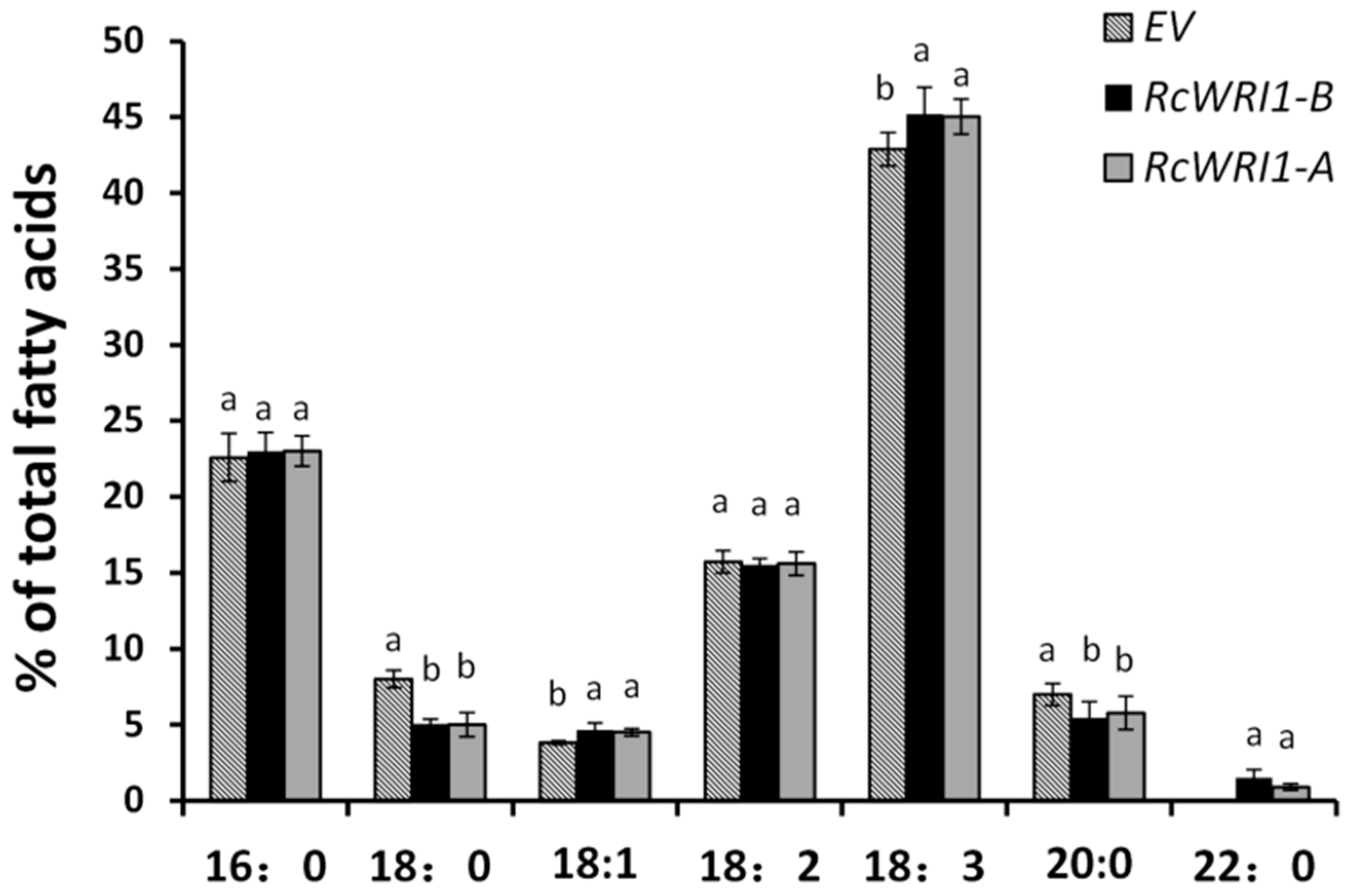
2.6. Ectopic Expression of RcWRI1s Changes Fatty Acid Profiles in N. benthamiana Leaves
3. Discussion
3.1. Castor RcWRI1 Expresses Two Alternative Splice Forms with Tissue-Specific Patterns
3.2. The Motif “VYL” Encoded by a 9-bp Exon Is an Important Component of WRI1 Proteins, But Not Necessary for Function of All WRI1s
3.3. Castor RcWRI1-A and RcWRI1-B Function to Increase Oil Biosynthesis in Heterogenous Tobacco Leaves by up Regulating the Target Genes Involved in Glycolysis and FA Synthesis
4. Materials and Method
4.1. Plant Materials
4.2. Castor Bean RcWRI1 Cloning
4.3. Sequence Alignment and Phylogenetic Analysis
4.4. Vector Construction
4.5. Overexpressing RcWRI1s in the Arabidopsis wri1-1 Mutant
4.6. Transient Expression in Tobacco Leaves
4.7. Real-Time PCR to Examine Expression of RcWRI1-A and RcWRI1-B
4.8. Lipid Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author contributions
Conflicts of Interest
Abbreviations
| TAGs | triacylglycerols |
| ORF | open reading frame |
| PKp-β1 | plastid pyruvate kinase beta subunit 1 |
| PDH-E1α | dehydrogenase E1 component alpha subunit |
| BCCP2 | biotin carboxyl carrier protein isoform 2 |
| ACCase | acetyl-CoA carboxylase |
| SUS2 | sucrose synthase 2 |
| KAS1 | 3-ketoacyl-acyl carrier protein synthase 1 |
| Pyr | pyruvate |
| CoA | acetyl-coenzyme A |
| ER | endoplasmic reticulum |
| G3P | glycerol-3-phosphate |
| DAF | days after florescence |
| FFAP | free fatty acid phase |
| FAME | FA methyl esters |
| MS | Murashige and Skoog |
| PPT | phosphinothricin |
| MES | MES monohydrate |
| FW | fresh weigh |
| EST | expressed sequence tag |
References
- Vanhercke, T.; El Tahchy, A.; Liu, Q.; Zhou, X.R.; Shrestha, P.; Divi, U.K.; Ral, J.P.; Mansour, M.P.; Nichols, P.D.; James, C.N.; et al. Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 2014, 12, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Durrett, T.P.; Benning, C.; Ohlrogge, J. Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 2008, 54, 593–607. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Napier, J.A.; Clemente, T.E.; Cahoon, E.B. New frontiers in oilseed biotechnology: Meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr. Opin. Biotechnol. 2011, 22, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Lepiniec, L. Physiological and developmental regulation of seed oil production. Prog. Lipid Res. 2010, 49, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Stymne, S.; Ohlrogge, J. Biochemical pathways in seed oil synthesis. Curr. Opin. Plant Biol. 2013, 16, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Li, R.; Hildebrand, D.F. Biosynthesis and metabolic engineering of palmitoleate production, an important contributor to human health and sustainable industry. Prog. Lipid Res. 2012, 51, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, X.; Chang, M.; Gao, Y.; Zhang, L.; Xue, J.A.; Run-Zhi, L.I. New type of industrial oilseed crop Camelina Sativa: From genome to metabolic engineering. Plant Physiol. J. 2015, 51, 1204–1216. [Google Scholar]
- Weselake, R.J.; Taylor, D.C.; Rahman, M.H.; Shah, S.; Laroche, A.; McVetty, P.B.; Harwood, J.L. Increasing the flow of carbon into seed oil. Biotechnol. Adv. 2009, 27, 866–878. [Google Scholar] [CrossRef] [PubMed]
- Napier, J.A. The production of unusual fatty acids in transgenic plants. Annu. Rev. Plant Biol. 2007, 58, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Bates, P.D.; Durrett, T.P.; Ohlrogge, J.B.; Pollard, M. Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 2009, 150, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Cernac, A.; Benning, C. Wrinkled1 Encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 2004, 40, 575–585. [Google Scholar] [CrossRef] [PubMed]
- Focks, N.; Benning, C. Wrinkled1: A novel, low-seed-oil mutant of arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998, 118, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Baud, S.; Bourrellier, A.B.F.; Azzopardi, M.; Berger, A.; Dechorgnat, J.; Daniel-Vedele, F.; Lepiniec, L.; Miquel, M.; Rochat, C.; Hodges, M.; et al. Pii Is Induced by wrinkled1 and fine-tunes fatty acid composition in seeds of Arabidopsis thaliana. Plant J. 2010, 64, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, N.D.; Bates, P.D.; Browse, J. Wrinkled1 Rescues Feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physiol. 2016, 171, 179–191. [Google Scholar] [CrossRef] [PubMed]
- To, A.; Joubes, J.; Barthole, G.; Lecureuil, A.; Scagnelli, A.; Jasinski, S.; Lepiniec, L.; Baud, S. Wrinkled transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 2012, 24, 5007–5023. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.T.; Lu, X.; Song, Q.X.; Chen, H.W.; Wei, W.; Tao, J.J.; Bian, X.H.; Shen, M.; Ma, B.; Zhang, W.K.; et al. Selection for a zinc-finger protein contributes to seed oil increase during soybean domestication. Plant Physiol. 2017, 173, 2208–2224. [Google Scholar] [CrossRef] [PubMed]
- Maeo, K.; Tokuda, T.; Ayame, A.; Mitsui, N.; Kawai, T.; Tsukagoshi, H.; Ishiguro, S.; Nakamura, K. An AP2-type transcription factor, wrinkled1, of arabidopsis thaliana binds to the aw-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J. 2009, 60, 476–487. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hua, W.; Zhan, G.; Wei, F.; Wang, X.; Liu, G.; Wang, H. Increasing seed mass and oil content in transgenic arabidopsis by the overexpression of Wri1-Like gene from Brassica napus. Plant Physiol. Biochem. 2010, 48, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Allen, W.B.; Zheng, P.; Li, C.; Glassman, K.; Ranch, J.; Nubel, D.; Tarczynski, M.C. Expression of Zmlec1 and Zmwri1 Increases seed oil production in Maize. Plant Physiol. 2010, 153, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Dahee, A.; Kim, H.; Ju, S.; Go, Y.S.; Kim, H.U.; Suh, M.C. Expression of Camelina wrinkled1 isoforms rescue the seed phenotype of the Arabidopsis Wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 2017, 8, 34. [Google Scholar]
- Bourgis, F.; Kilaru, A.; Cao, X.; Ngando-Ebongue, G.F.; Drira, N.; Ohlrogge, J.B.; Arondel, V. Comparative Transcriptome and metabolite analysis of oil palm and date palm mesocarp that differ dramatically in carbon partitioning. Proc. Natl. Acad. Sci. USA 2011, 108, 12527–12532. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kong, Q.; Arondel, V.; Kilaru, A.; Bates, P.D.; Thrower, N.A.; Benning, C.; Ohlrogge, J.B. Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 2013, 8, e68887. [Google Scholar] [CrossRef] [PubMed]
- Sjodin, A.; Street, N.R.; Sandberg, G.; Gustafsson, P.; Jansson, S. The populus genome integrative explorer (popgenie): A new resource for exploring the populus genome. New Phytol. 2009, 182, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Munz, J.; Cass, C.; Zienkiewicz, A.; Kong, Q.; Ma, W.; Sanjaya Sedbrook, J.; Benning, C. Ectopic expression of Wrinkled1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol. 2015, 169, 1836–1847. [Google Scholar] [PubMed]
- Grimberg, A.; Carlsson, A.S.; Marttila, S.; Bhalerao, R.; Hofvander, P. Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 2015, 15, 192. [Google Scholar] [CrossRef] [PubMed]
- Akande, T.O.; Odunsi, A.A.; Akinfala, E.O. A review of nutritional and toxicological implications of Castor Bean (Ricinus Communis L.) meal in animal feeding systems. J. Anim. Physiol. Anim. Nutr. 2016, 100, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, A.; Guerci, A. Various uses of the castor oil plant (Ricinus communis L.). A Review. J. Ethnopharmacol. 1982, 5, 117–137. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, L.; Tian, X.; Di, J.; Su, Y.; Huang, F.; Chen, Y. Crucial enzymes in the hydroxylated triacylglycerol-ricinoleate biosynthesis pathway of castor bean. Curr. Protein Pept. Sci. 2014, 15, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Ast, G. How did alternative splicing evolve? Nat. Rev. Genet. 2004, 5, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Priest, H.D.; Givan, S.A.; Shen, R.; Bryant, D.W.; Fox, S.E.; Wong, W.K.; Mockler, T.C. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010, 20, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, M.J.; Lim, M.H.; Kim, S.G.; Lee, M.; Baldwin, I.T.; Park, C.M. A self-regulatory circuit of circadian clock-associated1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar] [CrossRef] [PubMed]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; van Montagu, M.; Jofuku, K.D. The AP2 Domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar] [PubMed]
- Kim, H.U.; Lee, K.R.; Jung, S.J.; Shin, H.A.; Go, Y.S.; Suh, M.C.; Kim, J.B. Senescence-inducible LEC2 enhances triacylglycerol accumulation in leaves without negatively affecting plant growth. Plant Biotechnol. J. 2015, 13, 1346–1359. [Google Scholar] [CrossRef] [PubMed]
- Sanjaya, T.; Durrett, P.; Weise, S.E.; Benning, C. Increasing the energy density of vegetative tissues by diverting carbon from starch to oil biosynthesis in Transgenic Arabidopsis. Plant Biotechnol. J. 2011, 9, 874–883. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Yu, K.; Wu, Y.; Tateno, M.; Hatanaka, T.; Hildebrand, D.F. Vernonia DGATs can complement the disrupted oil and protein metabolism in epoxygenase-expressing soybean seeds. Metab. Eng. 2012, 14, 29–38. [Google Scholar] [CrossRef] [PubMed]




| Primer Name | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) |
|---|---|---|
| NbActin | CAGTGGCCGTACAACAGGTA | AACCGAAGAATTGCATGAGG |
| NbPKp-β1 | CTGTGTCGCTACGGACTGAA | GTGTTGGACATCATCGTTGC |
| NbPKp-α | TAAAAGCTCGGGGCATGGTC | GGTTTCCCAGCTAAGAACTTGT |
| NbPDH-E1α | TGGTAACGATGCTCTTGCTG | CTCCACCATTCGATTTCGTT |
| NbACP1 | GCAAATGGATCGAGGCTAAC | CTTGGAAGGATCGACTTTGG |
| NbKAS1 | CACCCATCAATTCACCAATG | CCCATCCCAGTTATGACCAC |
| NbBCCP2 | GCTGATTCGTCTGGAACCAT | GCCTTCCACCTATGTTGCAT |
| RcActin | GTGCTTGATTCTGGTGATGGC | TTGGCAGTCTCAAGTTCTTGCTC |
| RcWRI1-A | GAAGGGAAGACAAGTTTATTTGG | ATTCAAGGTTGTCTCTGGTCC |
| RcWRI1-B | AAGGGAAGACAAGGGGCCTATG | GCTTTGGCGTCGAAGAGATG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, X.-J.; Mao, X.; Hao, Q.-T.; Liu, B.-L.; Xue, J.-A.; Li, R.-Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. Int. J. Mol. Sci. 2018, 19, 146. https://doi.org/10.3390/ijms19010146
Ji X-J, Mao X, Hao Q-T, Liu B-L, Xue J-A, Li R-Z. Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves. International Journal of Molecular Sciences. 2018; 19(1):146. https://doi.org/10.3390/ijms19010146
Chicago/Turabian StyleJi, Xia-Jie, Xue Mao, Qing-Ting Hao, Bao-Ling Liu, Jin-Ai Xue, and Run-Zhi Li. 2018. "Splice Variants of the Castor WRI1 Gene Upregulate Fatty Acid and Oil Biosynthesis When Expressed in Tobacco Leaves" International Journal of Molecular Sciences 19, no. 1: 146. https://doi.org/10.3390/ijms19010146