Alignment of Mitotic Chromosomes in Human Cells Involves SR-Like Splicing Factors Btf and TRAP150
Abstract
:1. Introduction
2. Results
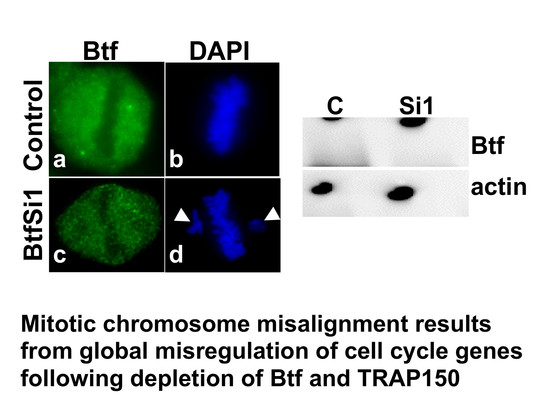
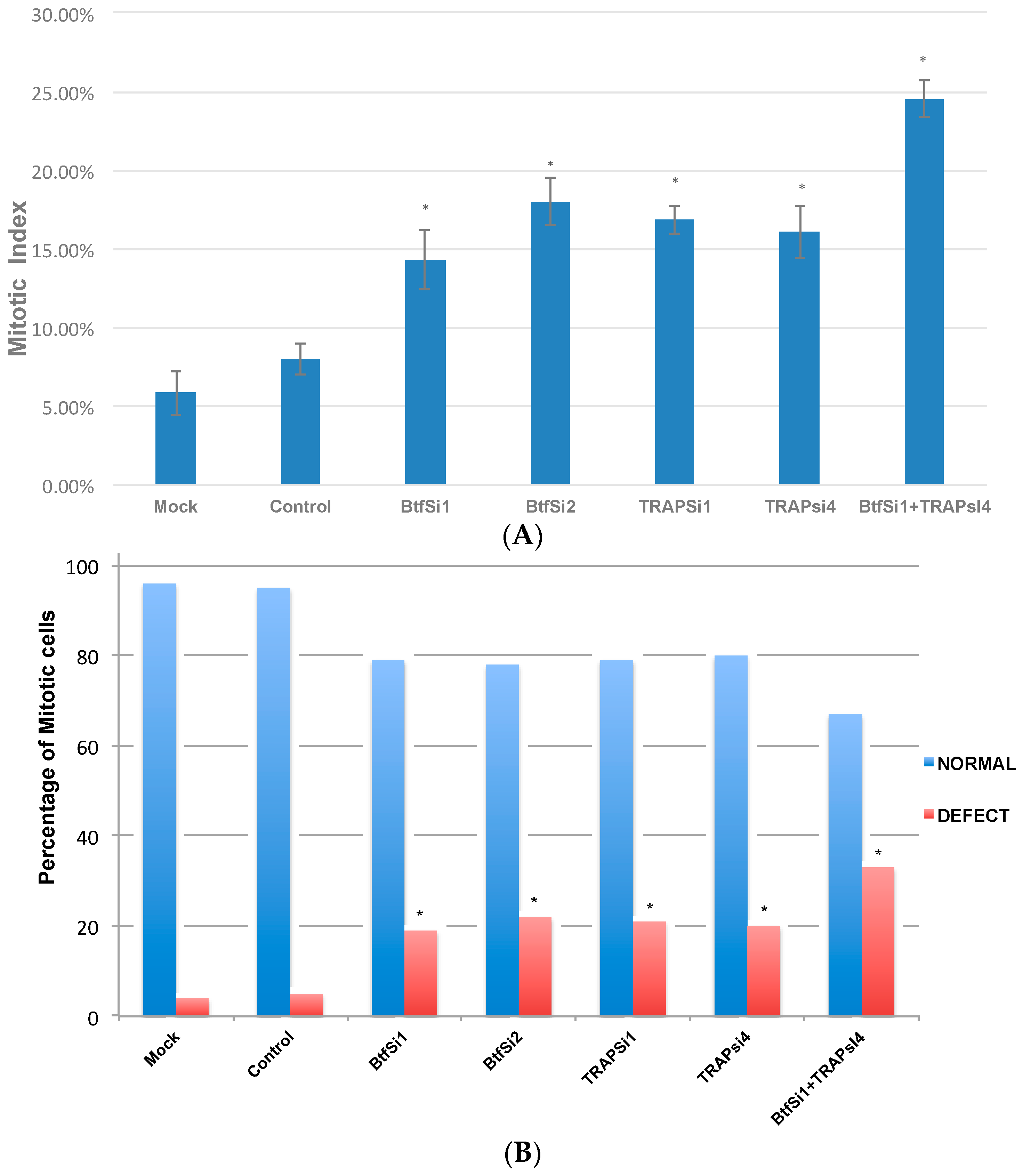
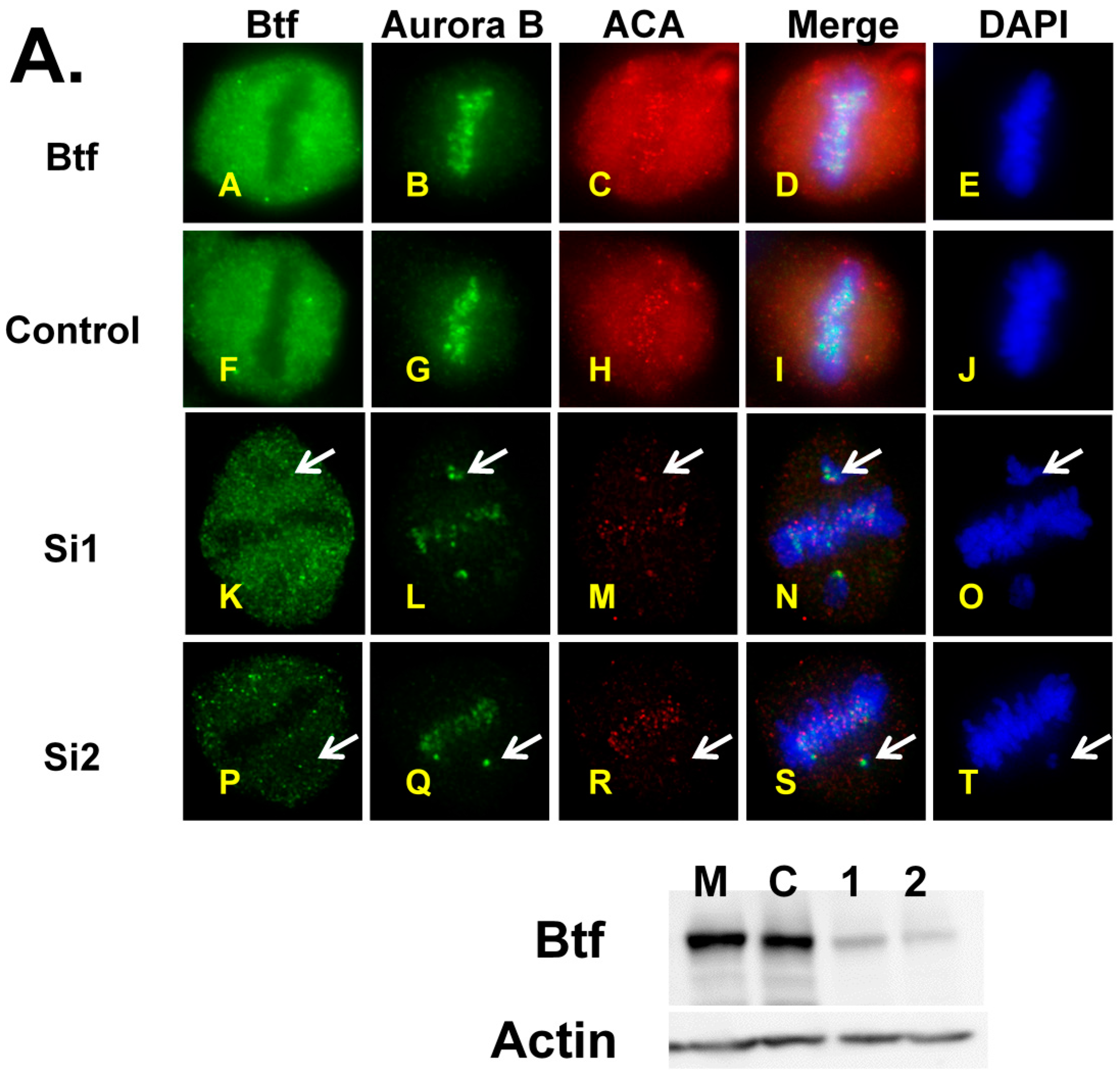
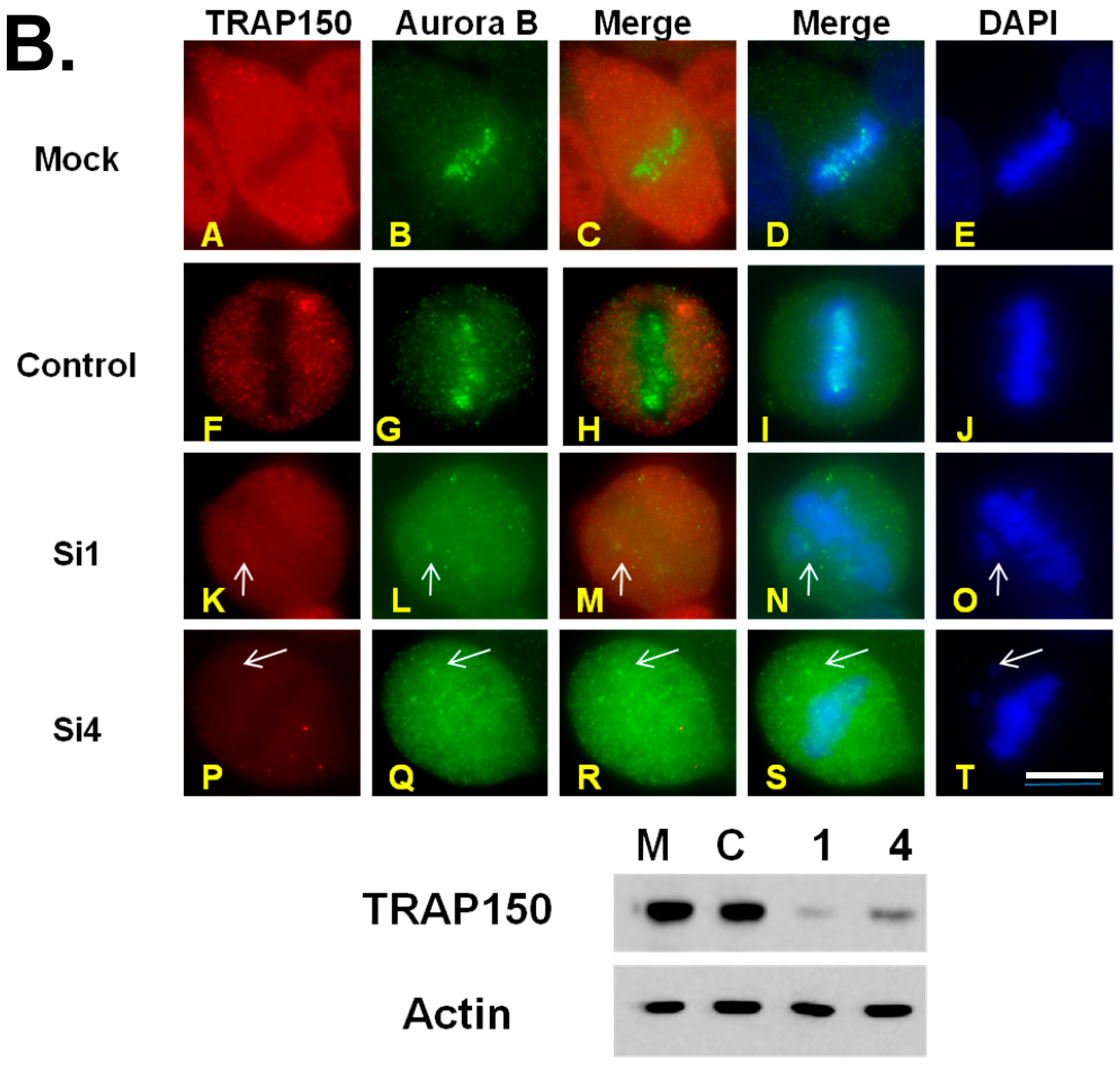
2.1. Depletion of Btf and/or TRAP150 Results in Mitotic Defects
2.2. Btf and TRAP150 Regulate Cell Cycle Progression in HeLa Cells
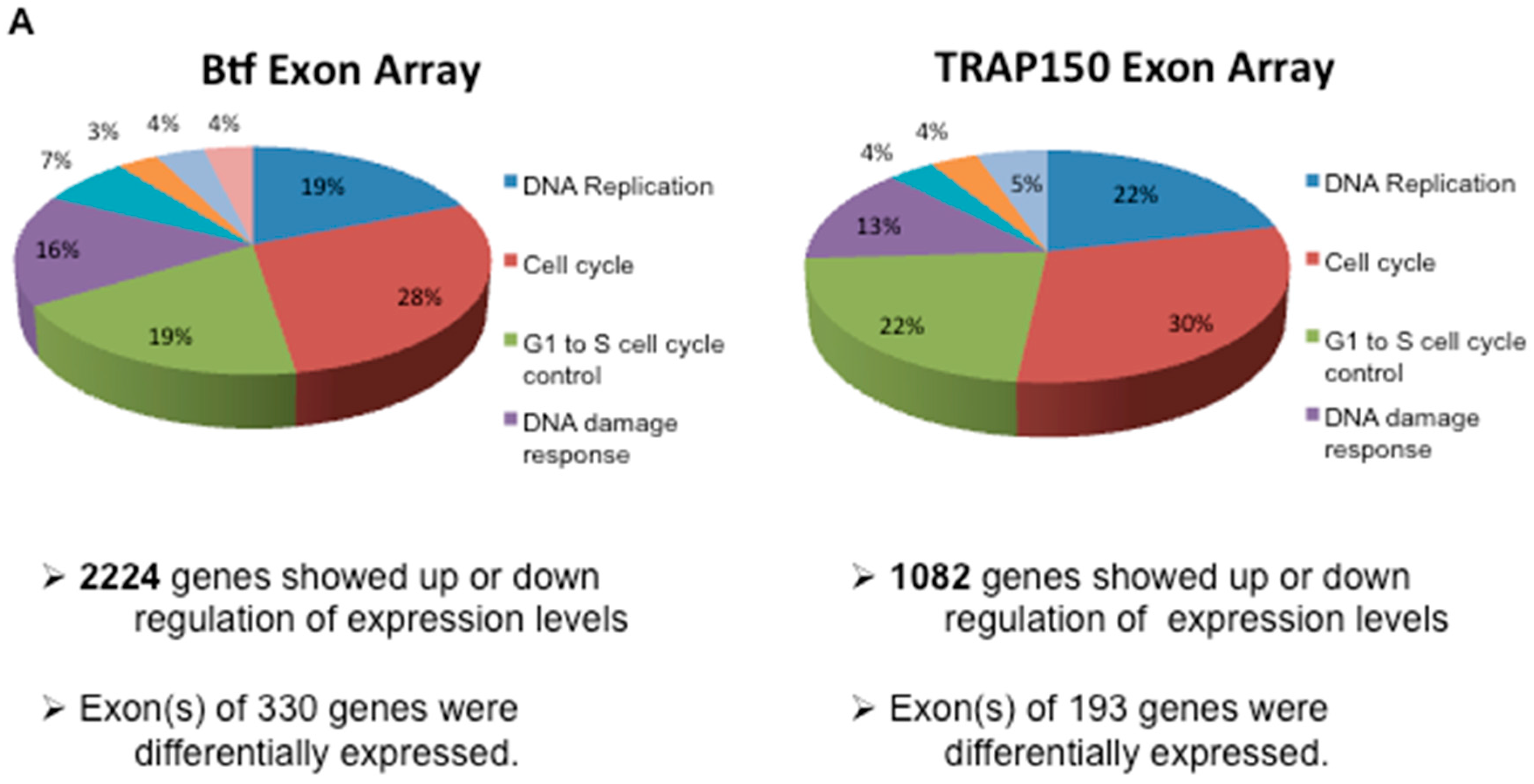
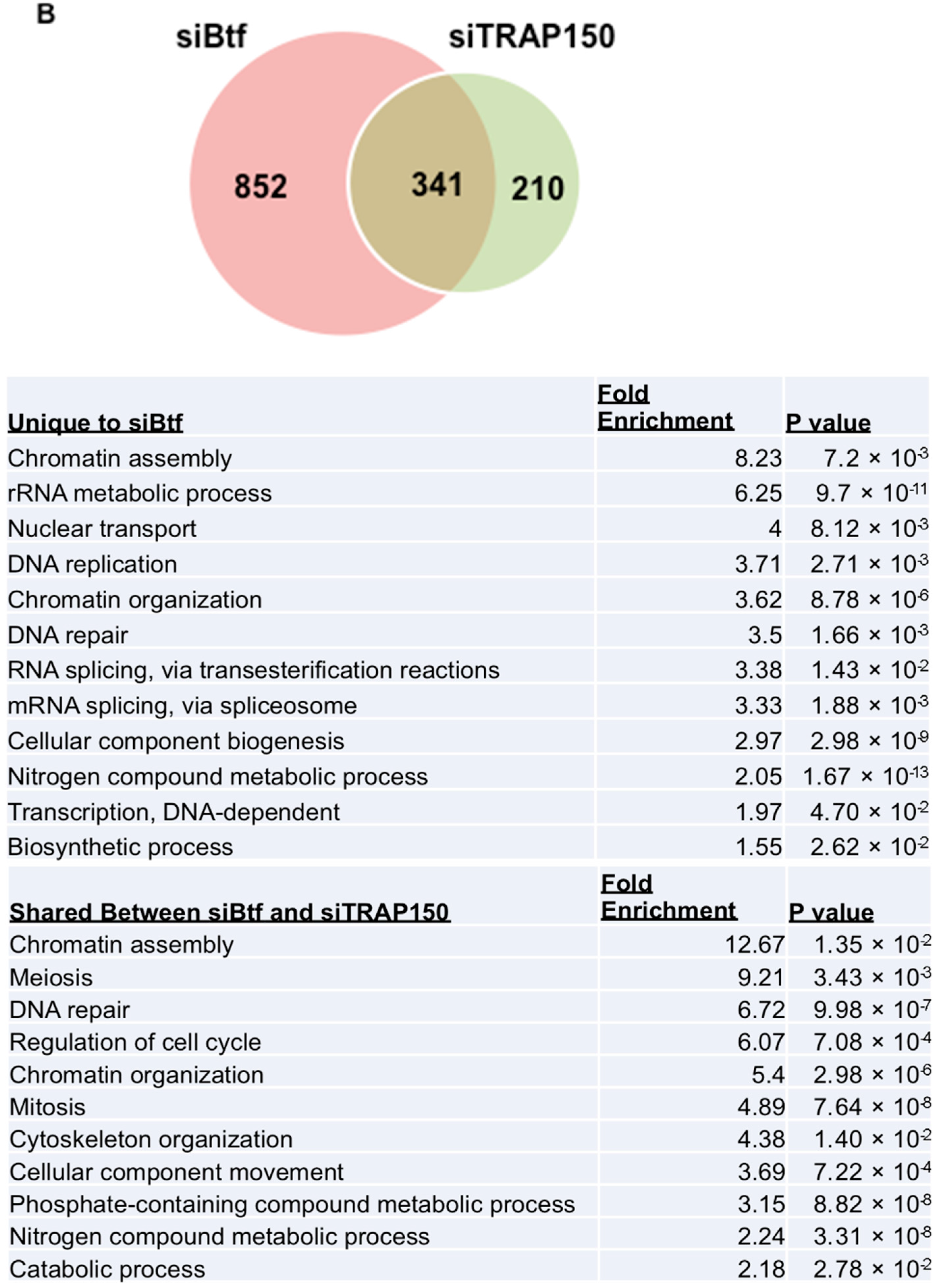
2.3. Btf and/or TRAP150 Depletion Alters Expression of Cell Cycle Regulator Transcripts
2.4. Mitotic Checkpoint Protein Aurora-B Localizes to Misaligned Chromosomes at Metaphase in Btf- and/or TRAP150-Depleted Cells
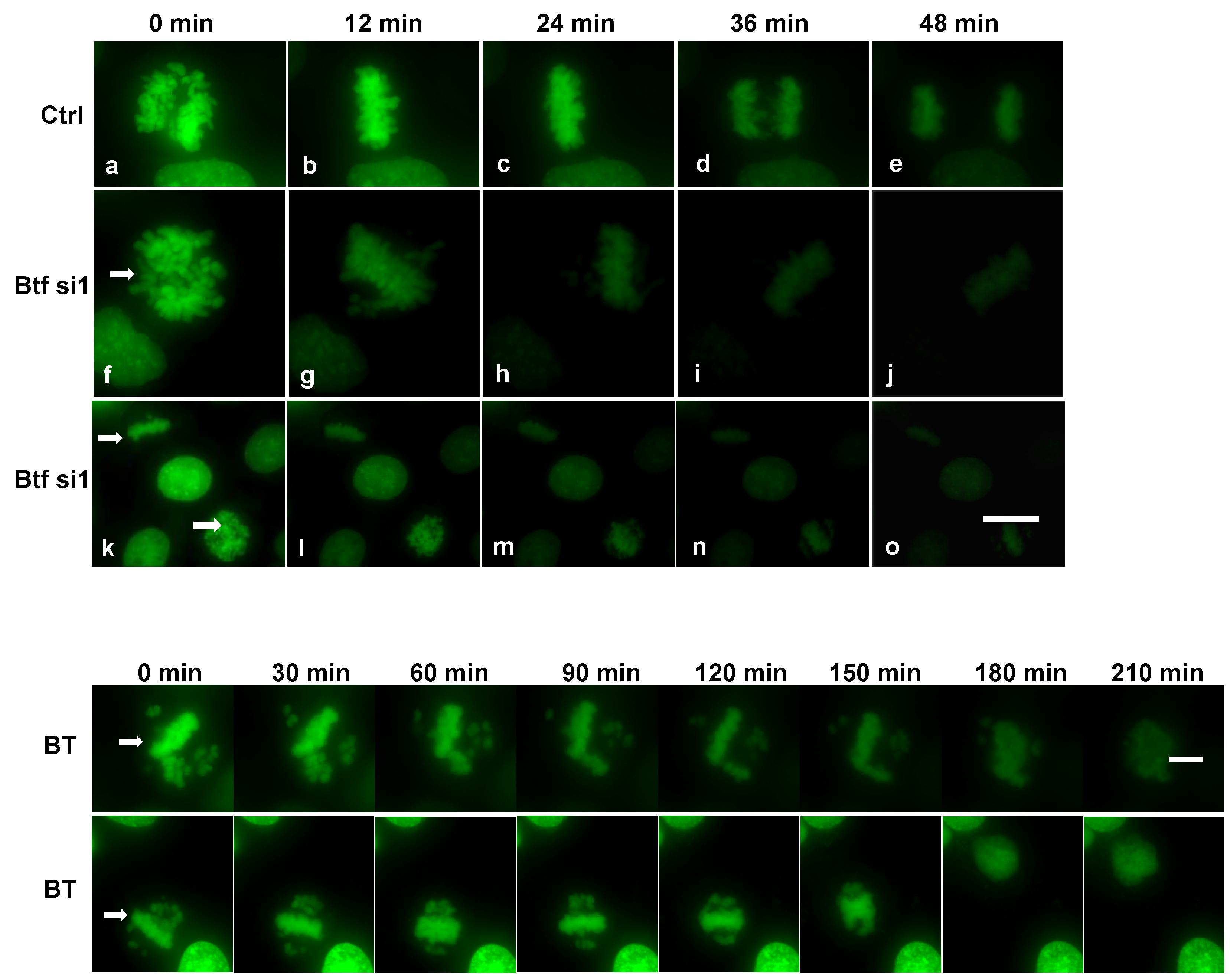
2.5. Mitosis Is Delayed or Incomplete in the Absence of Btf and/or TRAP150
3. Discussion
4. Materials and Methods
4.1. Cell Culture and RNAi
4.2. Immunofluorescence
4.3. Cell Extract Preparation and Immunoblotting
4.4. Flow Cytometry Analysis
4.5. RNA Extraction and DNase Treatment
4.6. Quantitative RT-PCR
4.7. Global Transcript Analysis
4.8. Live Cell Microscopy
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Btf | Bcl-2 associated transcription factor-1; also called BCLAF |
| TRAP150 | Thyroid hormone receptor-associated protein 150; also called THRAP3 |
| SR protein | Serine-arginine rich protein |
References
- Spector, D.L.; Landon, S.; O’Keefe, R.T. Organization of RNA polymerase II transcription and pre-mRNA splicing within the mammalian cell nucleus. Biochem. Soc. Trans. 1993, 21, 918–920. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, N.; Spahr, C.S.; Patterson, S.D.; Bubulya, P.; Neuwald, A.F.; Spector, D.L. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 2004, 15, 3876–3890. [Google Scholar] [CrossRef] [PubMed]
- Mintz, P.J.; Patterson, S.D.; Neuwald, A.F.; Spahr, C.S.; Spector, D.L. Purification and biochemical characterization of interchromatin granule clusters. EMBO J. 1999, 18, 4308–4320. [Google Scholar] [CrossRef] [PubMed]
- Manley, J.L.; Krainer, A.R. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010, 24, 1073–1194. [Google Scholar] [CrossRef] [PubMed]
- Kasof, G.M.; Goyal, L.; White, E. Btf, a novel death-promoting transcriptional repressor that interacts with Bcl-2-related proteins. Mol. Cell. Biol. 1999, 19, 4390–4404. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Yuan, C.X.; Malik, S.; Gu, W.; Fondell, J.D.; Yamamura, S.; Fu, Z.Y.; Zhang, X.; Qin, J.; Roeder, R.G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol. Cell 1999, 3, 361–370. [Google Scholar] [CrossRef]
- Rachez, C.; Freedman, L.P. Mediator complexes and transcription. Curr. Opin. Cell Biol. 2001, 13, 274–280. [Google Scholar] [CrossRef]
- Varia, S.; Potabathula, D.; Deng, Z.; Bubulya, A.; Bubulya, P.A. Btf and TRAP150 have distinct roles in regulating subcellular mRNA distribution. Nucleus 2013, 4, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Kittler, R.; Putz, G.; Pelletier, L.; Poser, I.; Heninger, A.K.; Drechsel, D.; Fischer, S.; Konstantinova, I.; Habermann, B.; Grabner, H.; et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature 2004, 432, 1036–1040. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. New talents for an old acquaintance: The SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle 2005, 4, 1706–1708. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Sun, Y.; Ding, J.H.; Lin, S.; Rose, D.W.; Rosenfeld, M.G.; Fu, X.D.; Li, X. Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell Biol. 2007, 27, 5393–5402. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nicklas, R.B. Mitotic forces control a cell-cycle checkpoint. Nature 1995, 373, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Schultz, A.; Cole, R.; Sluder, G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 1994, 127, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.B.; Pearson, C.G.; Yen, T.J.; Howell, B.J.; Salmon, E.D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 2001, 12, 1995–2009. [Google Scholar] [CrossRef] [PubMed]
- Howell, B.J.; Moree, B.; Farrar, E.M.; Stewart, S.; Fang, G.; Salmon, E.D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004, 14, 953–964. [Google Scholar] [CrossRef] [PubMed]
- Fang, G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell 2002, 13, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Bharadwaj, R.; Li, B.; Yu, H. Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell 2001, 1, 227–237. [Google Scholar] [CrossRef]
- Musacchio, A.; Hardwick, K.G. The spindle checkpoint: Structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Huaiyu, M.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2016, 45. [Google Scholar] [CrossRef]
- Sharma, A.; Markey, M.; Torres-Muñoz, K.; Varia, S.; Kadakia, M.; Bubulya, A.; Bubulya, P.A. Son maintains accurate splicing for a subset of human pre-mRNAs. J. Cell Sci. 2011, 124, 4286–4298. [Google Scholar] [CrossRef] [PubMed]
- Emig, D.; Salomonis, N.; Baumbach, J.; Lengauer, T.; Conklin, B.R.; Albrecht, M. AltAnalyze and DomainGraph: Analyzing and visualizing exon expression data. Nucleic Acids Res. 2010, 38, W755–W762. [Google Scholar] [CrossRef] [PubMed]


| Transcript Name | Btf Si1 | Btf Si2 | TRAPSi4 | |||
|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value | |
| AURK-B | 2.5 | 0.0002 | 1.7 | 0.003 | 2 | 0.001 |
| CENP-E | 2.8 | 0.003 | 2.5 | 0.027 | 3.0 | 0.012 |
| CENP-F | 4.4 | 0.0002 | 3.4 | 0.0015 | 4.1 | 0.01 |
| BUB1 | 3.36 | 0.002 | 3.16 | 0.005 | 3.38 | 0.002 |
| BUB1B | 5.46 | 0.0012 | 3.3 | 0.003 | 3.5 | 0.002 |
| CDK1 | 2.3 | 0.004 | 2.15 | 0.004 | 4.69 | 0.02 |
| CDC-20 | 3.38 | 0.003 | 2.37 | 0.0002 | 5.8 | 0.02 |
| MAD2L1 | 4.3 | 0.002 | 2.14 | 0.001 | 3.12 | 0.004 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varia, S.; Cheedu, D.; Markey, M.; Torres-Shafer, K.; Battini, V.P.; Bubulya, A.; Bubulya, P.A. Alignment of Mitotic Chromosomes in Human Cells Involves SR-Like Splicing Factors Btf and TRAP150. Int. J. Mol. Sci. 2017, 18, 1956. https://doi.org/10.3390/ijms18091956
Varia S, Cheedu D, Markey M, Torres-Shafer K, Battini VP, Bubulya A, Bubulya PA. Alignment of Mitotic Chromosomes in Human Cells Involves SR-Like Splicing Factors Btf and TRAP150. International Journal of Molecular Sciences. 2017; 18(9):1956. https://doi.org/10.3390/ijms18091956
Chicago/Turabian StyleVaria, Sapna, Divya Cheedu, Michael Markey, Keshia Torres-Shafer, Vishnu Priya Battini, Athanasios Bubulya, and Paula A. Bubulya. 2017. "Alignment of Mitotic Chromosomes in Human Cells Involves SR-Like Splicing Factors Btf and TRAP150" International Journal of Molecular Sciences 18, no. 9: 1956. https://doi.org/10.3390/ijms18091956