Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution
Abstract
:1. Introduction
2. General Characterization of Photoconvertible Fluorescent Proteins (pcFPs)
3. Least Evolved Ancestor (LEA) Series of pcFPs Developed by Ancestral Gene Reconstruction Technology
4. The pcFP LEA Is More Dynamic than Its Non-pcFP Precursor Protein
5. pcFPs Harbor a Softer, More Dynamic Active Site and a Remote Knob-Like Region That Is Highly Rigidified
6. The Charge States of Buried Functional Groups Appear to Control the Rate of Color Change
7. Transient Reverse Protonation of His193-Glu211 May Facilitate Deformation of the Chromophore in the Electronically Excited State
8. Functional Group Activation and Active Site Geometry Support a Concerted, One-Step Proton Abstraction and β-Elimination Reaction
9. LEA Is Photoconversion-Competent Because of Functional, Epistatic and Compensatory Substitutions
10. Concluding Remarks
10.1. Protein Dynamics and Protein Evolution
10.2. Design of Improved pcFPs for Super-Resolution Microscopy
Conflicts of Interest
Abbreviations
| ALL-GFP | Common green ancestor |
| LEA | Least evolved ancestor |
| avGFP | Aequorea victoria green fluorescent protein |
| dfi | Differential flexibility index |
| ESPT | Excited-state proton transfer |
| PRS | Perturbation response scanning |
| ENM | Elastic network model |
| pKaapp | Apparent acid dissociation constant |
References
- Lelimousin, M.; Noirclerc-Savoye, M.; Lazareno-Saez, C.; Paetzold, B.; le Vot, S.; Chazal, R.; Macheboeuf, P.; Field, M.J.; Bourgeois, D.; Royant, A. Intrinsic dynamics in ECFP and cerulean control fluorescence quantum yield. Biochemistry 2009, 48, 10038–10046. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Frontiera, R.R.; Tran, R.; Mathies, R.A. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature 2009, 462, 200. [Google Scholar] [CrossRef] [PubMed]
- Henderson, J.N.; Ai, H.W.; Campbell, R.E.; Remington, S.J. Structural basis for reversible photobleaching of a green fluorescent protein homologue. Proc. Natl. Acad. Sci. USA 2007, 104, 6672–6677. [Google Scholar] [CrossRef] [PubMed]
- Andresen, M.; Stiel, A.C.; Trowitzsch, S.; Weber, G.; Eggeling, C.; Wahl, M.C.; Hell, S.W.; Jakobs, S. Structural basis for reversible photoswitching in Dronpa. Proc. Natl. Acad. Sci. USA 2007, 104, 13005–13009. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Abulwerdi, F.; Baldridge, A.; Kowalik, J.; Solntsev, K.M.; Tolbert, L.M. Isomerization in Fluorescent Protein Chromophores Involves Addition/Elimination. J. Am. Chem. Soc. 2008, 130, 14096–14098. [Google Scholar] [CrossRef] [PubMed]
- Adam, V.; Lelimousin, M.; Boehme, S.; Desfonds, G.; Nienhaus, K.; Field, M.J.; Wiedenmann, J.; McSweeney, S.; Nienhaus, G.U.; Bourgeois, D. Structural characterization of IrisFP, an optical highlighter undergoing multiple photo-induced transformations. Proc. Natl. Acad. Sci. USA 2008, 105, 18343–18348. [Google Scholar] [CrossRef] [PubMed]
- Berardozzi, R.; Adam, V.; Martins, A.; Bourgeois, D. Arginine 66 Controls Dark-State Formation in Green-to-Red Photoconvertible Fluorescent Proteins. J. Am. Chem. Soc. 2016, 138, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Wachter, R.M.; Watkins, J.L.; Kim, H. Mechanistic diversity of red fluorescence acquisition by GFP-like proteins. Biochemistry 2010, 49, 7417–7427. [Google Scholar] [CrossRef] [PubMed]
- Ando, R.; Hama, H.; Yamamoto-Hino, M.; Mizuno, H.; Miyawaki, A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 12651–12656. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, J.; Ivanchenko, S.; Oswald, F.; Schmitt, F.; Roecker, C.; Salih, A.; Spindler, K.D.; Nienhaus, G.U. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 2004, 101, 15905–15910. [Google Scholar] [CrossRef] [PubMed]
- McEvoy, A.L.; Hoi, H.; Bates, M.; Platonova, E.; Cranfill, P.J.; Baird, M.A.; Davidson, M.W.; Ewers, H.; Liphardt, J.; Campbell, R.E. mMaple: A Photoconvertible Fluorescent Protein for Use in Multiple Imaging Modalities. PLoS ONE 2012, 7, e51314. [Google Scholar] [CrossRef] [PubMed]
- Wiedenmann, J.; Gayda, S.; Adam, V.; Oswald, F.; Nienhaus, K.; Bourgeois, D.; Nienhaus, G.U. From EosFP to mIrisFP: Structure-based development of advanced photoactivatable marker proteins of the GFP-family. J. Biophotonics 2011, 4, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Zou, T.; Modi, C.; Doerner, K.; Grunkemeyer, T.J.; Chen, L.; Fromme, R.; Matz, M.V.; Ozkan, S.B.; Wachter, R.M. A hinge migration mechanism unlocks the evolution of green-to-red photoconversion in GFP-like proteins. Structure 2015, 23, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Zewail, A.H. Femtochemistry: Ultrafast Dynamics of the Chemical Bond; World Scientific: Singapore, 1994. [Google Scholar]
- Frontiera, R.R.; Fang, C.; Dasgupta, J.; Mathies, R.A. Probing structural evolution along multidimensional reaction coordinates with femtosecond stimulated Raman spectroscopy. Phys. Chem. Chem. Phys. 2012, 14, 405–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Patel, H.N.; Lappe, J.W.; Wachter, R.M. Reaction progress of chromophore biogenesis in green fluorescent protein. J. Am. Chem. Soc. 2006, 128, 4766–4772. [Google Scholar] [CrossRef] [PubMed]
- Pouwels, L.J.; Zhang, L.; Chan, N.; Dorrestein, P.; Wachter, R.M. Kinetic isotope effect studies on the de novo rate of chromophore formation in fast- and slow-maturing GFP variants. Biochemistry 2008, 47, 10111–10122. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Mal, T.K.; Tong, K.I.; Ando, R.; Furuta, T.; Ikura, M.; Miyawaki, A. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol. Cell 2003, 12, 1051–1058. [Google Scholar] [CrossRef]
- Nienhaus, K.; Nienhaus, G.U.; Wiedenmann, J.; Nar, H. Structural basis for photo-induced protein cleavage and green-to-red conversion of fluorescent protein EosFP. Proc. Natl. Acad. Sci. USA 2005, 102, 9156–9159. [Google Scholar] [CrossRef] [PubMed]
- Fron, E.; Sliwa, M.; Adam, V.; Michiels, J.; Rocha, S.; Dedecker, P.; Hofkens, J.; Mizuno, H. Excited state dynamics of the photoconvertible fluorescent protein Kaede revealed by ultrafast spectroscopy. Photochem. Photobiol. Sci. 2014, 13, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Ugalde, J.A.; Chang, B.S.W.; Matz, M.V. Evolution of coral pigments recreated. Science 2004, 305, 1433. [Google Scholar] [CrossRef] [PubMed]
- Field, S.F.; Matz, M.V. Retracing evolution of red fluorescence in GFP-like proteins from Faviina corals. Mol. Biol. Evol. 2010, 27, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Field, S.F.; Bulina, M.Y.; Kelmanson, I.V.; Bielawski, J.P.; Matz, M.V. Adaptive evolution of multicolored fluorescent proteins in reef-building corals. J. Mol. Evol. 2006, 62, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Alieva, N.O.; Konzen, K.A.; Field, S.F.; Meleshkevitch, E.A.; Hunt, M.E.; Beltran-Ramirez, V.; Miller, D.J.; Wiedenmann, J.; Salih, A.; Matz, M.V. Diversity and evolution of coral fluorescent proteins. PLoS ONE 2008, 3, e2680. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Grunkemeyer, T.J.; Modi, C.; Chen, L.; Fromme, R.; Matz, M.V.; Wachter, R.M. Acid-base catalysis and crystal structures of a least-evolved ancestral GFP-like protein undergoing green-to-red photoconversion. Biochemistry 2013, 52, 8048–8059. [Google Scholar] [CrossRef] [PubMed]
- Habuchi, S.; Tsutsui, H.; Kochaniak, A.B.; Miyawaki, A.; van Oijen, A.M. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS ONE 2008, 3, e3944. [Google Scholar] [CrossRef] [PubMed]
- Hoi, H.; Shaner, N.C.; Davidson, M.W.; Cairo, C.W.; Wang, J.; Campbell, R.E. A monomeric photoconvertible fluorescent protein for imaging of dynamic protein localization. J. Mol. Biol. 2010, 401, 776–791. [Google Scholar] [CrossRef] [PubMed]
- Atilgan, C.; Atilgan, A.R. Perturbation-response scanning reveals ligand entry-exit mechanisms of ferric binding protein. PLoS Comput. Biol. 2009, 5, e1000544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atilgan, C.; Gerek, Z.N.; Ozkan, S.B.; Atilgan, A.R. Manipulation of conformational change in proteins by single-residue perturbations. Biophys. J. 2010, 99, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, I.; Mizuno, H.; Tong, K.I.; Furuta, T.; Tanaka, F.; Yoshimura, M.; Miyawaki, A.; Ikura, M. Crystallographic evidence for water-assisted photo-induced peptide cleavage in the stony coral fluorescent protein Kaede. J. Mol. Biol. 2007, 2007, 918–926. [Google Scholar] [CrossRef] [PubMed]
- Karplus, P.A.; Pearson, M.A.; Hausinger, R.P. 70 Years of crystalline urease: What have we learned? Acc. Chem. Res. 1997, 30, 330–337. [Google Scholar] [CrossRef]
- Tsutsui, H.; Karasawa, S.; Shimizu, H.; Nukina, N.; Miyawaki, A. Semi-rational engineering of a coral fluorescent protein into an efficient highlighter. EMBO Rep. 2005, 6, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Stiel, A.C.; Trowitzsch, S.; Weber, G.; Andresen, M.; Eggeling, C.; Hell, S.W.; Jakobs, S.; Wahl, M.C. 1.8 A bright-state structure of the reversibly switchable fluorescent protein Dronpa guides the generation of fast switching variants. Biochem. J. 2007, 402, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, H.; Mal, T.K.; Walchli, M.; Kikuchi, A.; Fukano, T.; Ando, R.; Jeyakanthan, J.; Taka, J.; Shiro, Y.; Ikura, M.; et al. Light-dependent regulation of structural flexibility in a photochromic fluorescent protein. Proc. Natl. Acad. Sci. USA 2008, 105, 9227–9232. [Google Scholar] [CrossRef] [PubMed]
- Warren, M.M.; Kaucikas, M.; Fitzpatrick, A.; Champion, P.; Timothy Sage, J.; van Thor, J.J. Ground-state proton transfer in the photoswitching reactions of the fluorescent protein Dronpa. Nat. Commun. 2013, 4, 1461. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, H.; Shimizu, H.; Mizuno, H.; Nukina, N.; Furuta, T.; Miyawaki, A. The E1 mechanism in photo-induced β-elimination reactions for green-to-red conversion of fluorescent proteins. Chem. Biol. 2009, 16, 1140–1147. [Google Scholar] [CrossRef] [PubMed]
- Bahar, I.; Lezon, T.R.; Yang, L.W.; Eyal, E. Global dynamics of proteins: Bridging between structure and function. Annu. Rev. Biophys. 2010, 39, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Bhabha, G.; Lee, J.; Ekiert, D.C.; Gam, J.; Wilson, I.A.; Dyson, H.J.; Benkovic, S.J.; Wright, P.E. A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis. Science 2011, 332, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Tawfik, D.S. Protein dynamism and evolvability. Science 2009, 324, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Glembo, T.J.; Thorpe, M.F.; Farrell, D.W.; Gerek, Z.N.; Ozkan, S.B. Collective dynamics differentiates functional divergence in protein evolution. PLoS Comput. Biol. 2012, 8, e1002428. [Google Scholar] [CrossRef] [PubMed]
- Bhabha, G.; Ekiert, D.C.; Jennewein, M.; Zmasek, C.M.; Tuttle, L.M.; Kroon, G.; Dyson, H.J.; Godzik, A.; Wilson, I.A.; Wright, P.E. Divergent evolution of protein conformational dynamics in dihydrofolate reductase. Nat. Struct. Mol. Biol. 2013, 20, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Gerek, Z.N.; Kumar, S.; Ozkan, S.B. Structural dynamics flexibility informs function and evolution at a proteome scale. Evol. Appl. 2013, 6, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ortlund, E.A.; Bridgham, J.T.; Redinbo, M.R.; Thornton, J.W. Crystal structure of an ancient protein: Evolution by conformational epistasis. Science 2007, 317, 1544–1548. [Google Scholar] [CrossRef] [PubMed]
- Patterson, G.H.; Davidson, M.W.; Manley, S.; Lippincott-Schwartz, J. Supperresolution imaging using single-molecule localization. Annu. Rev. Phys. Chem. 2010, 61, 345–367. [Google Scholar] [CrossRef] [PubMed]
- Moeyaert, B.; Bich, N.N.; de Zitter, E.; Rocha, S.; Clays, K.; Mizuno, H.; van Meervelt, L.; Hofkens, J.; Dedecker, P. Green-to-red photoconvertible Dronpa mutant for multimodal super-resolution fluorescence microscopy. ACS Nano 2014, 8, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, P.; van Engelenburg, S.B.; Lippincott-Schwartz, J. Superresolution imaging of biological systems using photoactivated localization microscopy. Chem. Rev. 2014, 114, 3189–3202. [Google Scholar] [CrossRef] [PubMed]
- Nienhaus, K.; Nienhaus, G.U. Where do we stand with super-resolution optical microscopy? J. Mol. Biol. 2016, 428, 308–322. [Google Scholar] [CrossRef] [PubMed]
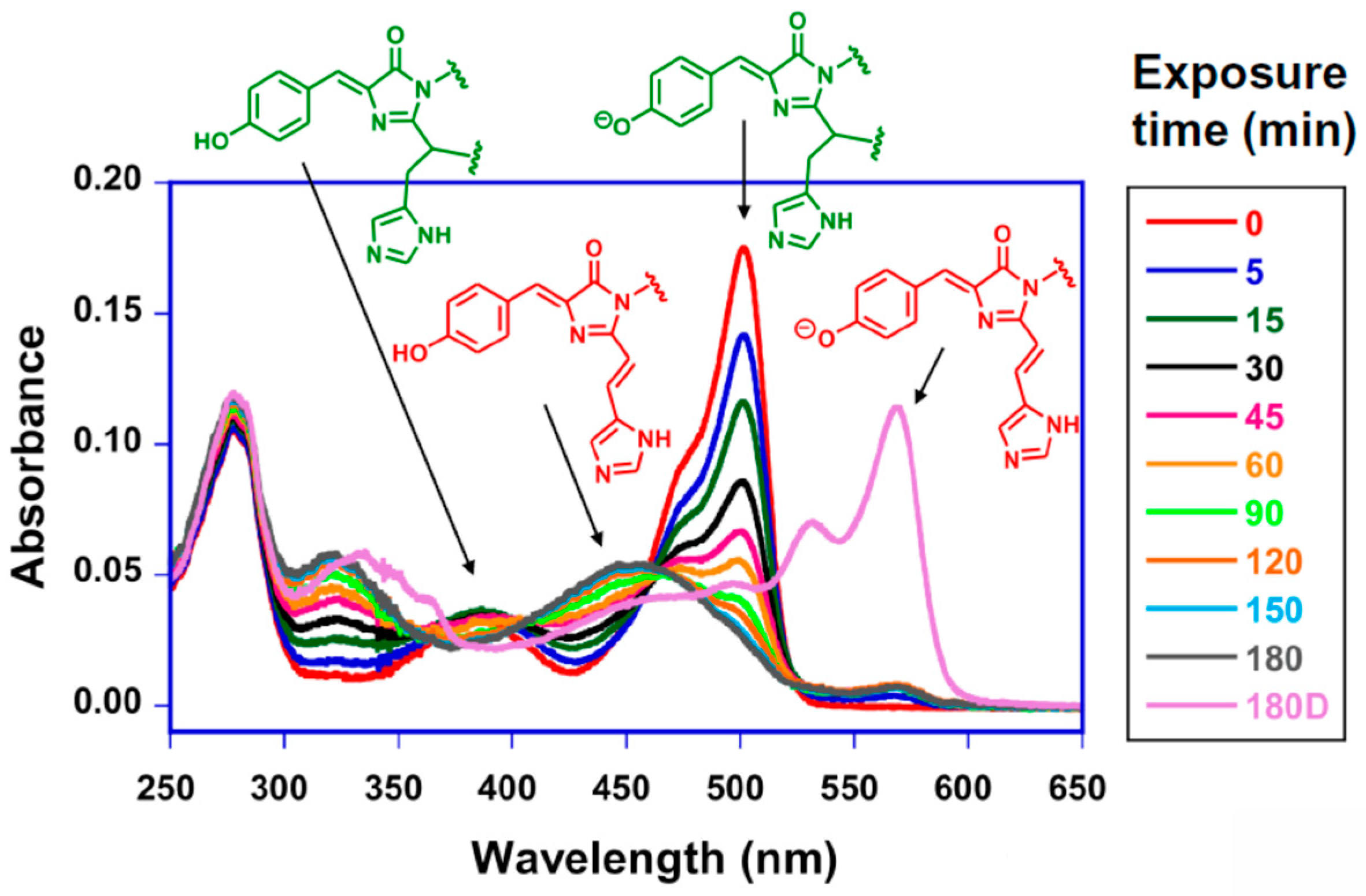
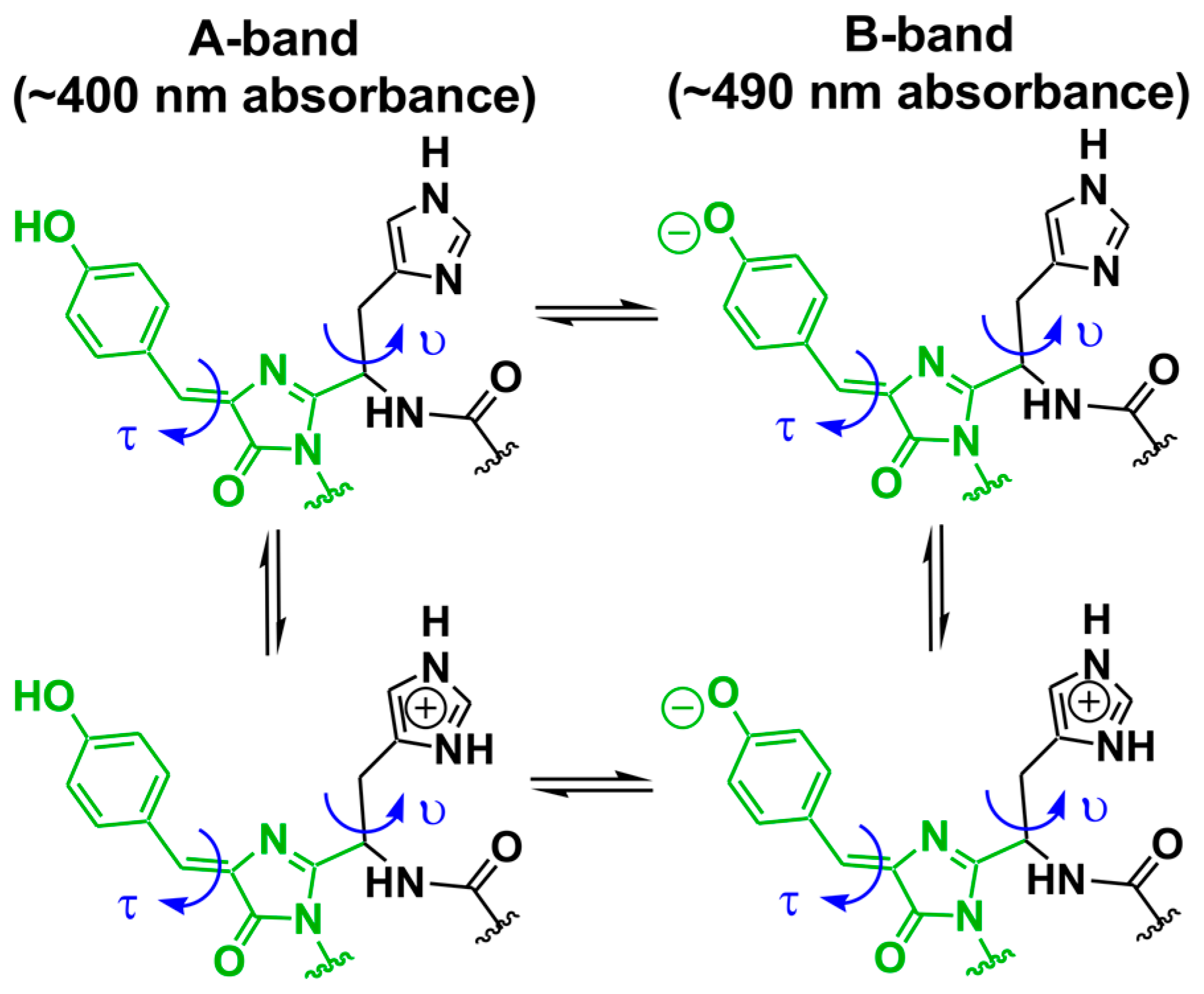
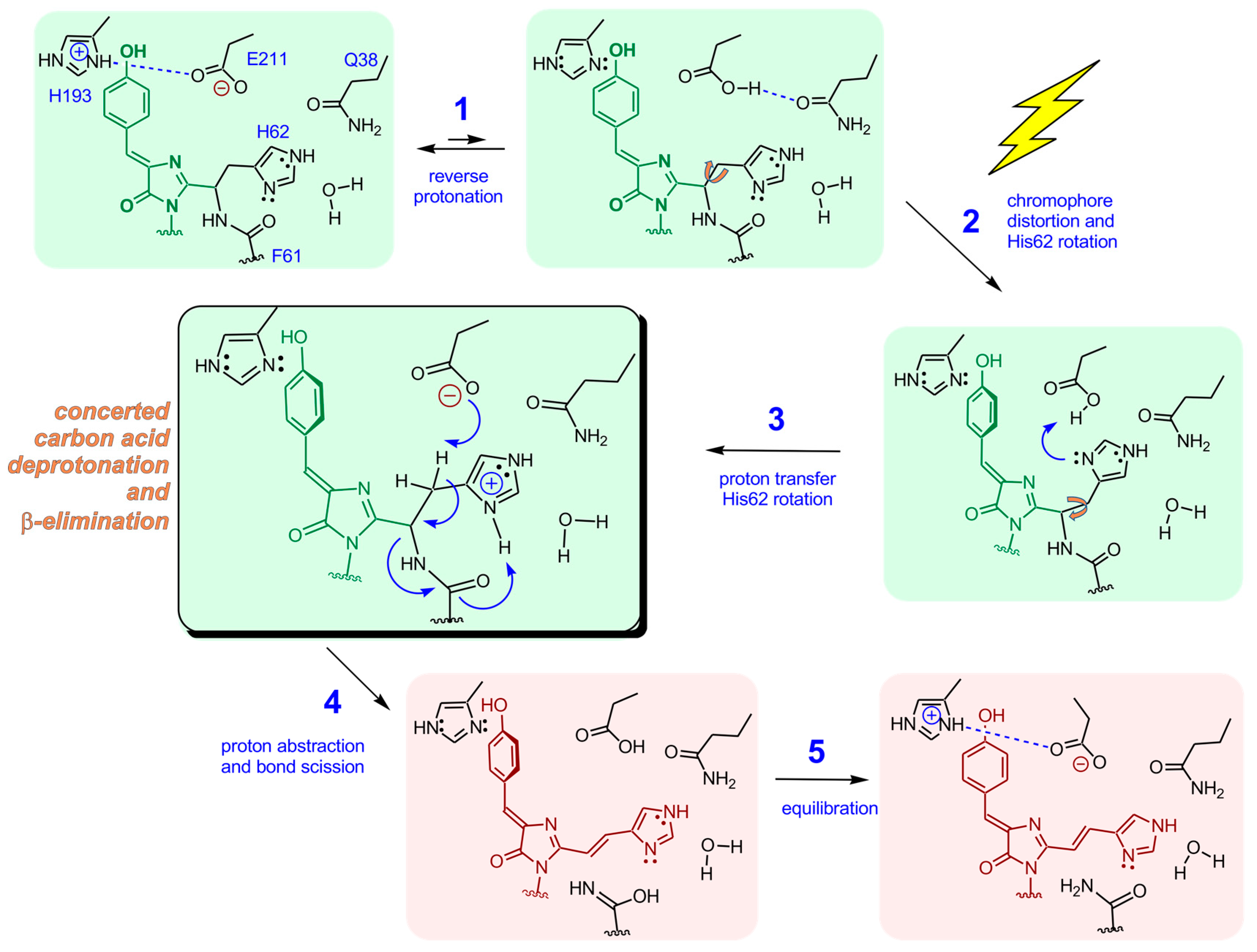
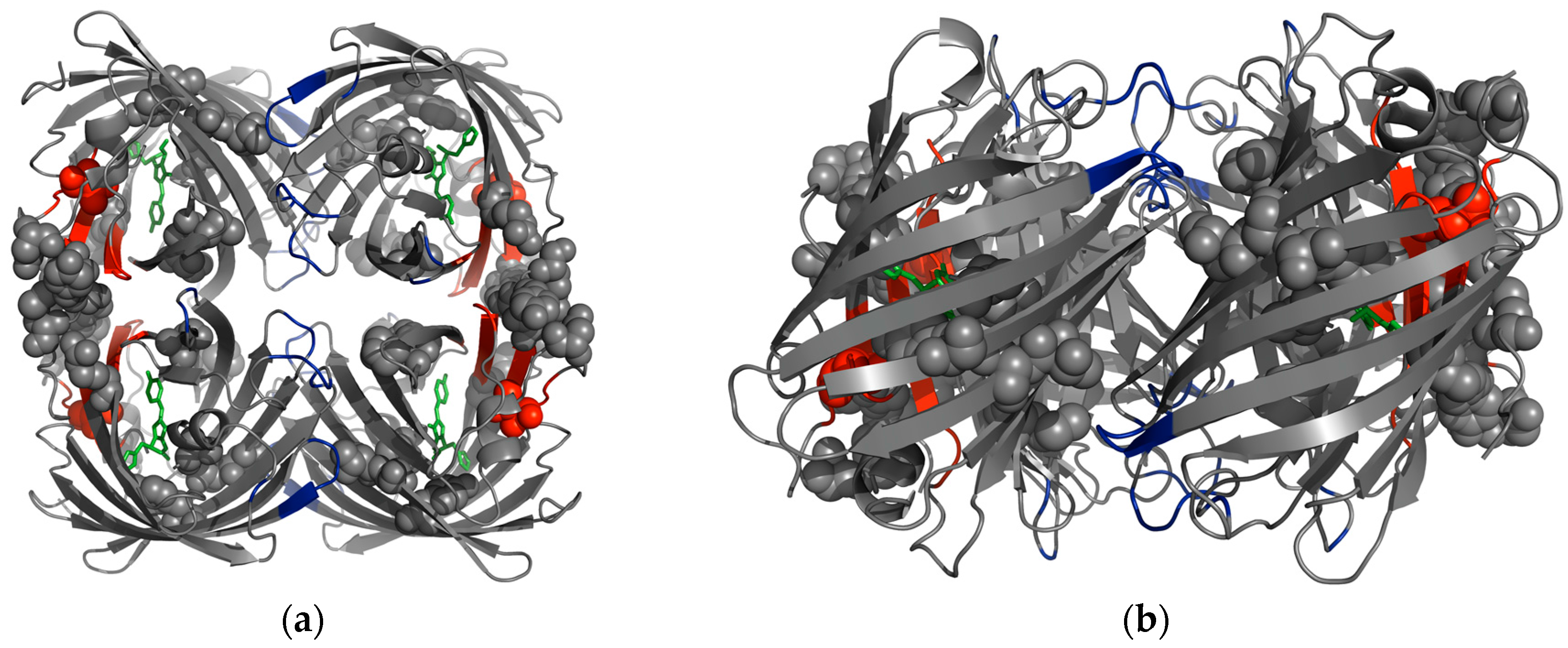
| Residue No. | ALL-GFP | LEA | ∆dfi | avGFP Residue No. |
|---|---|---|---|---|
| 60 | A | V | −0.03 | 63 |
| 62 | Q | H | +0.23 | 65 |
| 69 | T | A | +0.46 | 72 |
| 74 | D | H | −0.04 | 77 |
| 104 | T | R | −0.17 | 109 |
| 105 | S | N | +0.09 | 110 |
| 116 | Y | N | +0.05 | 121 |
| 154 | M | T | −0.04 | 162 |
| 157 | V | I | −0.06 | 165 |
| 194 | R | C | +0.23 | 204 |
| 216 | R | H | +0.15 | 227 |
| 217 | S | (deleted) | n/a | 228 * |
| 219 (218 for LEA) | L | G | n/a | 229 * |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wachter, R.M. Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution. Int. J. Mol. Sci. 2017, 18, 1792. https://doi.org/10.3390/ijms18081792
Wachter RM. Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution. International Journal of Molecular Sciences. 2017; 18(8):1792. https://doi.org/10.3390/ijms18081792
Chicago/Turabian StyleWachter, Rebekka M. 2017. "Photoconvertible Fluorescent Proteins and the Role of Dynamics in Protein Evolution" International Journal of Molecular Sciences 18, no. 8: 1792. https://doi.org/10.3390/ijms18081792





