Optogenetic Investigation of Arousal Circuits
Abstract
:1. Introduction
2. The Importance of Sleep
Defining Different Sleep-Wake States
- Quiet wakefulness—slower EEG frequencies such as α (8–14 Hz) and β (15–24 Hz) [6] and muscle activity observed in the EMG signal;
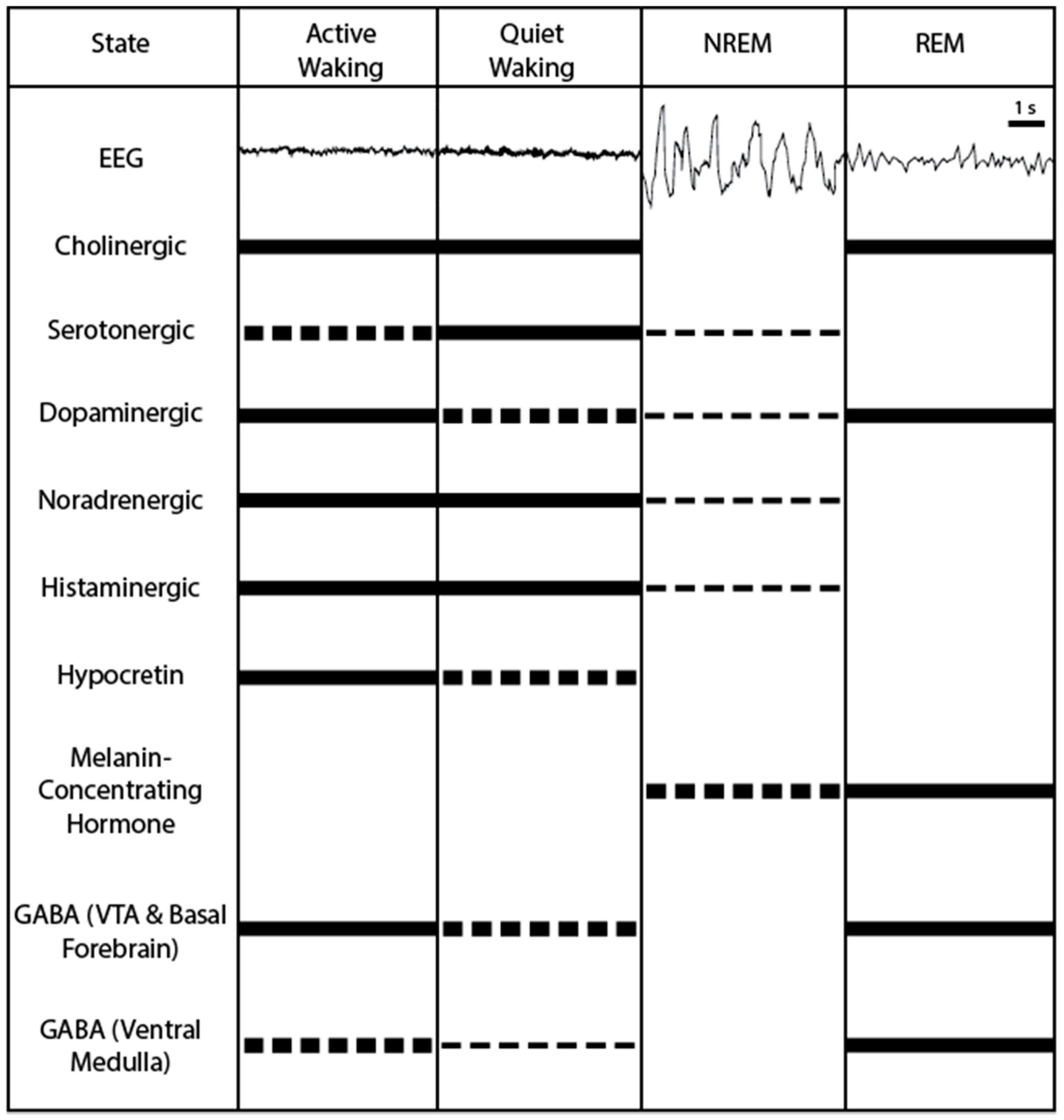
3. Neural Correlates of Sleep-Wake Modulation
3.1. Cholinergic Neurons: Active during Waking and REM Sleep
3.2. Serotonergic Neurons: Promoting Wakefulness
3.3. Noradrenergic Neurons: Promoting Wakefulness
3.4. Dopaminergic Neurons: Promoting Wakefulness
3.5. Neuropeptide S: Promoting Wakefulness
3.6. Hypocretin Neurons: Promoting Wakefulness
3.7. Melanin-Concentrating Hormone Neurons: Promoting REM Sleep
3.8. Glutamatergic and GABAergic Neurons
4. Optogenetic Investigation of Individual Neuron Populations
4.1. Hypocretin Neurons
4.2. Melanin-Concentrating Hormone Neurons
4.3. Dopaminergic Neurons
4.4. Cholinergic Neurons
4.5. GABAergic Neurons
4.6. Astrocytes
5. Optogenetic Tools for Circuit Investigation
5.1. Hypocretin Interactions with Histamine
5.2. Hypocretin Interactions with the Locus Coeruleus
5.3. Hypocretin Interactions with Leptin
5.4. Hypocretin Interactions with Melanin-Concentrating Hormone (Feat. GABA)
6. Further Developments and Future Directions
7. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| BdNST | Bed nucleus of the stria terminalis |
| CSF | Cerebrospinal fluid |
| DA | Dopaminergic |
| DREADD | Designer receptors exclusively activated by designer drugs |
| DRN | Dorsal raphe nucleus |
| EEG | Electroencephalogram |
| EMG | Electromyogram |
| GABA | γ Amino butyric acid |
| GFAP | Glial fibrillary acidic protein |
| Hcrt | Hypocretin |
| HcrtR2 | Hypocretin receptor 2 |
| Hz | Hertz |
| i.c.v. | Intracerebroventricular |
| LC | Locus coeruleus |
| LH | Lateral hypothalamus |
| MCH | Melanin-concentrating hormone |
| mRNA | Messenger ribonucleic acid |
| NE | Norepinephrine |
| NPS | Neuropeptide S |
| NREM | Non-rapid eye movement |
| REM | Rapid eye movement |
| TMN | Tuberomammillary nucleus |
| Vglut2 | Vesicular glutamate transporter 2 |
| VLPO | Ventrolateral pre-optic nucleus |
| vlTMN | Ventrolateral tuberomammillary nucleus |
| vPAG | Ventral periaqueductal gray |
| VTA | Ventral tegmental area |
References
- Aserinsky, E.; Kleitman, N. Regularly occurring periods of eye motility, and concomitant phenomena, during sleep. Science 1953, 118, 273–274. [Google Scholar] [CrossRef] [PubMed]
- Rechtschaffen, A.; Kales, A. A Manual of Standardized Terminology, Techniques, and Scoring Systems for Sleep Stages of Human Subjects; U.S. National Institute of Neurological Diseases and Blindness, Neurological Information Network: Maryland, MD, USA, 1969; Volum 26, p. 644.
- Huber, R.; Deboer, T.; Tobler, I. Prion protein: A role in sleep regulation? J. Sleep Res. 1999, 8, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Landolt, H.-P.; Rétey, J.V.; Tönz, K.; Gottselig, J.M.; Khatami, R.; Buckelmüller, I.; Achermann, P. Caffeine attenuates waking and sleep electroencephalographic markers of sleep homeostasis in humans. Neuropsychopharmacology 2004, 29, 1933. [Google Scholar] [CrossRef] [PubMed]
- Aeschbach, D.; Matthews, J.R.; Postolache, T.T.; Jackson, M.A.; Giesen, H.A.; Wehr, T.A. Two circadian rhythms in the human electroencephalogram during wakefulness. Am. J. Physiol. 1999, 277, R1771–R1779. [Google Scholar] [PubMed]
- Niedermeyer, E. The normal eeg of the waking adult. In Electroencephalography: Basic Principles, Clinical Applications, and Related Fields; Schomer, D.L., Lopes da Silva, F., Eds.; Lippincott Williams & Wilkins: Pennsyivania, PA, USA, 2005; Volum 9, pp. 155–164. [Google Scholar]
- Cirelli, C.; Tononi, G. Is sleep essential? PLoS Biol. 2008, 6, e216. [Google Scholar] [CrossRef] [PubMed]
- Franken, P.; Chollet, D.; Tafti, M. The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 2001, 21, 2610–2621. [Google Scholar] [PubMed]
- Vyazovskiy, V.V.; Ruijgrok, G.; Deboer, T.; Tobler, I. Running wheel accessibility affects the regional electroencephalogram during sleep in mice. Cereb. Cortex 2006, 16, 328. [Google Scholar] [CrossRef] [PubMed]
- Vyazovskiy, V.V.; Tobler, I. Regional differences in NREM sleep slow-wave activity in mice with congenital callosal dysgenesis. J. Sleep Res. 2005, 14, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Vyazovskiy, V.V.; Harris, K.D. Sleep and the single neuron: The role of global slow oscillations in individual cell rest. Nat. Rev. Neurosci. 2013, 14, 443. [Google Scholar] [CrossRef] [PubMed]
- Vyazovskiy, V.V.; Achermann, P.; Bo rbely, A.A.; Tobler, I. The dynamics of spindles and eeg slow-wave activity in NREM sleep in mice. Arch. Ital. Biol. 2004, 142, 511–523. [Google Scholar] [PubMed]
- Lüthi, A. Sleep spindles: Where they come from, what they do. Neuroscientist 2014, 20, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Nir, Y.; Staba, R.J.; Andrillon, T.; Vyazovskiy, V.V.; Cirelli, C.; Fried, I.; Tononi, G. Regional slow waves and spindles in human sleep. Neuron 2011, 70, 153–169. [Google Scholar] [CrossRef] [PubMed]
- Callaway, C.W.; Lydic, R.; Baghdoyan, H.A.; Hobson, J.A. Pontogeniculooccipital waves: Spontaneous visual system activity during rapid eye movement sleep. Cell. Mol. Neurobiol. 1987, 7, 105–149. [Google Scholar] [CrossRef] [PubMed]
- Datta, S. Cellular and chemical neuroscience of mammalian sleep. Sleep Med. 2010, 11, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Karashima, A.; Katayama, N.; Nakao, M. Enhancement of synchronization between hippocampal and amygdala θ waves associated with pontine wave density. J. Neurophysiol. 2010, 103, 2318–2325. [Google Scholar] [CrossRef] [PubMed]
- Siegel, J.M. Clues to the functions of mammalian sleep. Nature 2005, 437, 1264. [Google Scholar] [CrossRef] [PubMed]
- Economo, C.V. Sleep as a problem of localization. J. Nerv. Ment. Dis. 1930, 71, 249–259. [Google Scholar] [CrossRef]
- Peyron, C.; Faraco, J.; Rogers, W.; Ripley, B.; Overeem, S.; Charnay, Y.; Nevsimalova, S.; Aldrich, M.; Reynolds, D.; Albin, R.; et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat. Med. 2000, 6, 991–997. [Google Scholar] [PubMed]
- Thannickal, T.C.; Moore, R.Y.; Nienhuis, R.; Ramanathan, L.; Gulyani, S.; Aldrich, M.; Cornford, M.; Siegel, J.M. Reduced number of hypocretin neurons in human narcolepsy. Neuron 2000, 27, 469–474. [Google Scholar] [CrossRef]
- Rye, D.B. The two faces of eve: Dopamine’s modulation of wakefulness and sleep. Neurology 2004, 63, S2–S7. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.E.; Stevens, D.R.; Haas, H.L. The physiology of brain histamine. Prog. Neurobiol. 2001, 63, 637–672. [Google Scholar] [CrossRef]
- White, J.M.; Rumbold, G.R. Behavioural effects of histamine and its antagonists: A review. Psychopharmacology (Berl) 1988, 95, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.G.; Hassani, O.K.; Alonso, A.; Jones, B.E. Cholinergic basal forebrain neurons burst with θ during waking and paradoxical sleep. J. Neurosci. 2005, 25, 4365–4369. [Google Scholar] [CrossRef] [PubMed]
- Hassani, O.K.; Lee, M.G.; Henny, P.; Jones, B.E. Discharge profiles of identified GABAergic in comparison to cholinergic and putative glutamatergic basal forebrain neurons across the sleep–wake cycle. J. Neurosci. 2009, 29, 11828–11840. [Google Scholar] [CrossRef] [PubMed]
- Celesia, G.G.; Jasper, H.H. Acetylcholine released from cerebral cortex in relation to state of activation. Neurology 1966, 16, 1053–1063. [Google Scholar] [CrossRef] [PubMed]
- Jasper, H.H.; Tessier, J. Acetylcholine liberation from cerebral cortex during paradoxical (rem) sleep. Science 1971, 172, 601–602. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Comisarow, J.; Day, J.; Fibiger, H.C.; Reiner, P.B. State–dependent release of acetylcholine in rat thalamus measured by in vivo microdialysis. J. Neurosci. 1994, 14, 5236–5242. [Google Scholar] [PubMed]
- Xu, M.; Chung, S.; Zhang, S.; Zhong, P.; Ma, C.; Chang, W.C.; Weissbourd, B.; Sakai, N.; Luo, L.; Nishino, S. Basal forebrain circuit for sleep–wake control. Nat. Neurosci. 2015, 18, 1641–1647. [Google Scholar] [CrossRef] [PubMed]
- El Mansari, M.; Sakai, K.; Jouvet, M. Unitary characteristics of presumptive cholinergic tegmental neurons during the sleep–waking cycle in freely moving cats. Exp. Brain Res. 1989, 76, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Koyama, Y. Are there cholinergic and non-cholinergic paradoxical sleep-on neurones in the pons? Neuroreport 1996, 7, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Grant, S.J.; Highfield, D.A. Extracellular characteristics of putative cholinergic neurons in the rat laterodorsal tegmental nucleus. Brain Res. 1991, 559, 64–74. [Google Scholar] [CrossRef]
- Steriade, M.; Datta, S.; Pare, D.; Oakson, G.; Dossi, R.C.C. Neuronal activities in brain-stem cholinergic nuclei related to tonic activation processes in thalamocortical systems. J. Neurosci. 1990, 10, 2541–2559. [Google Scholar] [PubMed]
- Boucetta, S.; Cissé, Y.; Mainville, L.; Morales, M.; Jones, B.E. Discharge profiles across the sleep–waking cycle of identified cholinergic, GABAergic, and glutamatergic neurons in the pontomesencephalic tegmentum of the rat. J. Neurosci. 2014, 34, 4708–4727. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, B.L.; Fornal, C.A. Activity of brain serotonergic neurons in the behaving animal. Pharmacol. Rev. 1991, 43, 563–578. [Google Scholar] [PubMed]
- Wilkinson, L.O.; Auerbach, S.B.; Jacobs, B.L. Extracellular serotonin levels change with behavioral state but not with pyrogen-induced hyperthermia. J. Neurosci. 1991, 11, 2732–2741. [Google Scholar] [PubMed]
- Portas, C.M.; Bjorvatn, B.; Ursin, R. Serotonin and the sleep/wake cycle: Special emphasis on microdialysis studies. Prog. Neurobiol. 2000, 60, 13–35. [Google Scholar] [CrossRef]
- Dahan, L.; Astier, B.; Vautrelle, N.; Urbain, N.; Kocsis, B.; Chouvet, G. Prominent burst firing of dopaminergic neurons in the ventral tegmental area during paradoxical sleep. Neuropsychopharmacology 2007, 32, 1232–1241. [Google Scholar] [CrossRef] [PubMed]
- Eban–Rothschild, A.; Rothschild, G.; Giardino, W.J.; Jones, J.R.; de Lecea, L. Vta dopaminergic neurons regulate ethologically relevant sleep–wake behaviors. Nat. Neurosci. 2016, 19, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.; Morilak, D.A.; Jacobs, B.L. Single unit activity of locus coeruleus neurons in the freely moving cat. I. During naturalistic behaviors and in response to simple and complex stimuli. Brain Res. 1986, 371, 324–334. [Google Scholar] [CrossRef]
- Hobson, J.A.; McCarley, R.W.; Wyzinski, P.W. Sleep cycle oscillation: Reciprocal discharge by two brainstem neuronal groups. Science 1975, 189, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Saper, C.B.; Fuller, P.M.; Pedersen, N.P.; Lu, J.; Scammell, T.E. Sleep state switching. Neuron 2010, 68, 1023–1042. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E. Arousal systems. Front. Biosci. 2003, 8, s438–s451. [Google Scholar] [CrossRef] [PubMed]
- Vanni-Mercier, G.; Gigout, S.; Debilly, G.; Lin, J.S. Waking selective neurons in the posterior hypothalamus and their response to histamine H3-receptor ligands: An electrophysiological study in freely moving cats. Behav. Brain Res. 2003, 144, 227–241. [Google Scholar] [CrossRef]
- John, J.; Wu, M.F.; Boehmer, L.N.; Siegel, J.M. Cataplexy-active neurons in the hypothalamus: Implications for the role of histamine in sleep and waking behavior. Neuron 2004, 42, 619–634. [Google Scholar] [CrossRef]
- Takahashi, K.; Lin, J.S.; Sakai, K. Neuronal activity of histaminergic tuberomammillary neurons during wake–sleep states in the mouse. J. Neurosci. 2006, 26, 10292–10298. [Google Scholar] [CrossRef] [PubMed]
- Estabrooke, I.V.; McCarthy, M.T.; Ko, E.; Chou, T.C.; Chemelli, R.M.; Yanagisawa, M.; Saper, C.B.; Scammell, T.E. Fos expression in orexin neurons varies with behavioral state. J. Neurosci. 2001, 21, 1656–1662. [Google Scholar] [PubMed]
- Modirrousta, M.; Mainville, L.; Jones, B.E. Orexin and MCH neurons express c-Fos differently after sleep deprivation vs. recovery and bear different adrenergic receptors. Eur. J. Neurosci. 2005, 21, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Kiyashchenko, L.I.; Mileykovskiy, B.Y.; Maidment, N.; Lam, H.A.; Wu, M.F.; John, J.; Peever, J.; Siegel, J.M. Release of hypocretin (orexin) during waking and sleep states. J. Neurosci. 2002, 22, 5282–5286. [Google Scholar] [PubMed]
- Alam, M.N.; Gong, H.; Alam, T.; Jaganath, R.; McGinty, D.; Szymusiak, R. Sleep–waking discharge patterns of neurons recorded in the rat perifornical lateral hypothalamic area. J. Physiol. 2002, 538, 619–631. [Google Scholar] [CrossRef] [PubMed]
- Koyama, Y.; Takahashi, K.; Kodama, T.; Kayama, Y. State-dependent activity of neurons in the perifornical hypothalamic area during sleep and waking. Neuroscience 2003, 119, 1209–1219. [Google Scholar] [CrossRef]
- Lee, M.G.; Hassani, O.K.; Jones, B.E. Discharge of identified orexin/hypocretin neurons across the sleep–waking cycle. J. Neurosci. 2005, 25, 6716–6720. [Google Scholar] [CrossRef] [PubMed]
- Mileykovskiy, B.Y.; Kiyashchenko, L.I.; Siegel, J.M. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron 2005, 46, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Lin, J.S.; Sakai, K. Neuronal activity of orexin and non-orexin waking-active neurons during wake–sleep states in the mouse. Neuroscience 2008, 153, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Zeitzer, J.M.; Buckmaster, C.L.; Parker, K.J.; Hauck, C.M.; Lyons, D.M.; Mignot, E. Circadian and homeostatic regulation of hypocretin in a primate model: Implications for the consolidation of wakefulness. J. Neurosci. 2003, 23, 3555–3560. [Google Scholar] [PubMed]
- Hanriot, L.; Camargo, N.; Courau, A.C.; Leger, L.; Luppi, P.H.; Peyron, C. Characterization of the melanin-concentrating hormone neurons activated during paradoxical sleep hypersomnia in rats. J. Comp. Neurol. 2007, 505, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Verret, L.; Goutagny, R.; Fort, P.; Cagnon, L.; Salvert, D.; Leger, L.; Boissard, R.; Salin, P.; Peyron, C.; Luppi, P.H. A role of melanin-concentrating hormone producing neurons in the central regulation of paradoxical sleep. BMC Neurosci. 2003, 4, 19. [Google Scholar] [CrossRef] [PubMed]
- Hassani, O.K.; Lee, M.G.; Jones, B.E. Melanin-concentrating hormone neurons discharge in a reciprocal manner to orexin neurons across the sleep–wake cycle. Proc. Natl. Acad. Sci. USA 2009, 106, 2418–2422. [Google Scholar] [CrossRef] [PubMed]
- Blouin, A.M.; Fried, I.; Wilson, C.L.; Staba, R.J.; Behnke, E.J.; Lam, H.A.; Maidment, N.T.; Karlsson, K.A.E.; Lapierre, J.L.; Siegel, J.M. Human hypocretin and melanin-concentrating hormone levels are linked to emotion and social interaction. Nat. Commun. 2013, 4, 1547. [Google Scholar] [CrossRef] [PubMed]
- Steffensen, S.C.; Lee, R.S.; Stobbs, S.H.; Henriksen, S.J. Responses of ventral tegmental area GABA neurons to brain stimulation reward. Brain Res. 2001, 906, 190–197. [Google Scholar] [CrossRef]
- Weber, F.; Chung, S.; Beier, K.T.; Xu, M.; Luo, L.; Dan, Y. Control of REM sleep by ventral medulla GABAergic neurons. Nature 2015, 526, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.M.; Saper, C.B.; Levey, A.I.; Wainer, B.H.; Terry, R.D. Distribution of cholinergic neurons in rat brain: Demonstrated by the immunocytochemical localization of choline acetyltransferase. J. Comp. Neurol. 1983, 216, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Mesulam, M.M.; Mufson, E.J.; Wainer, B.H.; Levey, A.I. Central cholinergic pathways in the rat: An overview based on an alternative nomenclature (ch1–ch6). Neuroscience 1983, 10, 1185–1201. [Google Scholar] [CrossRef]
- Duque, A.; Balatoni, B.; Detari, L.; Zaborszky, L. EEG correlation of the discharge properties of identified neurons in the basal forebrain. J. Neurophysiol. 2000, 84, 1627–1635. [Google Scholar] [PubMed]
- Manns, I.D.; Alonso, A.; Jones, B.E. Rhythmically discharging basal forebrain units comprise cholinergic, GABAergic, and putative glutamatergic cells. J. Neurophysiol. 2003, 89, 1057–1066. [Google Scholar] [CrossRef] [PubMed]
- Boucetta, S.; Jones, B.E. Activity profiles of cholinergic and intermingled GABAergic and putative glutamatergic neurons in the pontomesencephalic tegmentum of urethane-anesthetized rats. J. Neurosci. 2009, 29, 4664–4674. [Google Scholar] [CrossRef] [PubMed]
- Fisahn, A.; Pike, F.G.; Buhl, E.H.; Paulsen, O. Cholinergic induction of network oscillations at 40 Hz in the hippocampus in vitro. Nature 1998, 394, 186–189. [Google Scholar] [CrossRef] [PubMed]
- Huerta, P.T.; Lisman, J.E. Heightened synaptic plasticity of hippocampal ca1 neurons during a cholinergically induced rhythmic state. Nature 1993, 364, 723–725. [Google Scholar] [CrossRef] [PubMed]
- Shimono, K.; Brucher, F.; Granger, R.; Lynch, G.; Taketani, M. Origins and distribution of cholinergically induced β rhythms in hippocampal slices. J. Neurosci. 2000, 20, 8462–8473. [Google Scholar] [PubMed]
- Berntson, G.G.; Shafi, R.; Sarter, M. Specific contributions of the basal forebrain corticopetal cholinergic system to electroencephalographic activity and sleep/waking behaviour. Eur. J. Neurosci. 2002, 16, 2453–2461. [Google Scholar] [CrossRef] [PubMed]
- Kapas, L.; Obal, F., Jr.; Book, A.A.; Schweitzer, J.B.; Wiley, R.G.; Krueger, J.M. The effects of immunolesions of nerve growth factor-receptive neurons by 192 IgG-saporin on sleep. Brain Res. 1996, 712, 53–59. [Google Scholar] [CrossRef]
- Blanco-Centurion, C.; Xu, M.; Murillo–Rodriguez, E.; Gerashchenko, D.; Shiromani, A.M.; Salin-Pascual, R.J.; Hof, P.R.; Shiromani, P.J. Adenosine and sleep homeostasis in the basal forebrain. J. Neurosci. 2006, 26, 8092–8100. [Google Scholar] [CrossRef] [PubMed]
- Kalinchuk, A.V.; McCarley, R.W.; Stenberg, D.; Porkka-Heiskanen, T.; Basheer, R. The role of cholinergic basal forebrain neurons in adenosine–mediated homeostatic control of sleep: Lessons from 1 to 92 IgG-saporin lesions. Neuroscience 2008, 157, 238–253. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Junek, A.; Black, M.A.; Semba, K. Effects of ibotenate and 192 IgG-saporin lesions of the nucleus basalis magnocellularis/substantia innominata on spontaneous sleep and wake states and on recovery sleep after sleep deprivation in rats. J. Neurosci. 2008, 28, 491–504. [Google Scholar] [CrossRef] [PubMed]
- Shiromani, P.J.; Fishbein, W. Continuous pontine cholinergic microinfusion via mini-pump induces sustained alterations in rapid eye movement (REM) sleep. Pharmacol. Biochem. Behav. 1986, 25, 1253–1261. [Google Scholar] [CrossRef]
- Gillin, J.C.; Wyatt, R.J.; Fram, D.; Snyder, F. The relationship between changes in REM sleep and clinical improvement in depressed patients treated with amitriptyline. Psychopharmacology 1978, 59, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Monti, J.M.; Jantos, H. The roles of dopamine and serotonin, and of their receptors, in regulating sleep and waking. Prog. Brain Res. 2008, 172, 625–646. [Google Scholar] [PubMed]
- Boutrel, B.; Monaca, C.; Hen, R.; Hamon, M.; Adrien, J. Involvement of 5-HT1A receptors in homeostatic and stress-induced adaptive regulations of paradoxical sleep: Studies in 5-HT1A knock-out mice. J. Neurosci. 2002, 22, 4686–4692. [Google Scholar] [PubMed]
- McGinty, D.J.; Harper, R.M. Dorsal raphe neurons: Depression of firing during sleep in cats. Brain Res. 1976, 101, 569–575. [Google Scholar] [CrossRef]
- Trulson, M.E.; Jacobs, B.L. Raphe unit activity in freely moving cats: Correlation with level of behavioral arousal. Brain Res. 1979, 163, 135–150. [Google Scholar] [CrossRef]
- Hammack, S.E.; Schmid, M.J.; LoPresti, M.L.; Der-Avakian, A.; Pellymounter, M.A.; Foster, A.C.; Watkins, L.R.; Maier, S.F. Corticotropin releasing hormone type 2 receptors in the dorsal raphe nucleus mediate the behavioral consequences of uncontrollable stress. J. Neurosci. 2003, 23, 1019–1025. [Google Scholar] [PubMed]
- Carlsson, A.; Falck, B.; Hillarp, N.A. Cellular localization of brain monoamines. Acta Physiol. Scand. Suppl. 1962, 56, 1–28. [Google Scholar] [PubMed]
- Flicker, C.; Geyer, M.A. The hippocampus as a possible site of action for increased locomotion during intracerebral infusions of norepinephrine. Behav. Neural. Biol. 1982, 34, 421–426. [Google Scholar] [CrossRef]
- Segal, D.S.; Mandell, A.J. Behavioral activation of rats during intraventricular infusion of norepinephrine. Proc. Natl. Acad. Sci. USA 1970, 66, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Harper, S.T.; Halaris, A.E. Effects of locus coeruleus lesions upon cerebral monoamine content, sleep–wakefulness states and the response to amphetamine in the cat. Brain Res. 1977, 124, 473–496. [Google Scholar] [CrossRef]
- Monti, J.M.; D’Angelo, L.; Jantos, H.; Barbeito, L.; Abo, V. Effect of DSP-4, a noradrenergic neurotoxin, on sleep and wakefulness and sensitivity to drugs acting on adrenergic receptors in the rat. Sleep 1988, 11, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Pompeiano, M.; Tononi, G. Neuronal gene expression in the waking state: A role for the locus coeruleus. Science 1996, 274, 1211–1215. [Google Scholar] [CrossRef] [PubMed]
- Cirelli, C.; Tononi, G. Differential expression of plasticity-related genes in waking and sleep and their regulation by the noradrenergic system. J. Neurosci. 2000, 20, 9187–9194. [Google Scholar] [PubMed]
- Cirelli, C.; Tononi, G. Locus ceruleus control of state-dependent gene expression. J. Neurosci. 2004, 24, 5410–5419. [Google Scholar] [CrossRef] [PubMed]
- Wisor, J.P.; Nishino, S.; Sora, I.; Uhl, G.H.; Mignot, E.; Edgar, D.M. Dopaminergic role in stimulant-induced wakefulness. J. Neurosci. 2001, 21, 1787–1794. [Google Scholar] [PubMed]
- Qu, W.-M.; Xu, X.-H.; Yan, M.-M.; Wang, Y.-Q.; Urade, Y.; Huang, Z.-L. Essential role of dopamine D2 receptor in the maintenance of wakefulness, but not in homeostatic regulation of sleep, in mice. J. Neurosci. 2010, 30, 4382–4389. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Jhou, T.C.; Saper, C.B. Identification of wake-active dopaminergic neurons in the ventral periaqueductal gray matter. J. Neurosci. 2006, 26, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.D.; Farber, J.; Gatz, P.; Roffwarg, H.; German, D.C. Activity of mesencephalic dopamine and non-dopamine neurons across stages of sleep and waking in the rat. Brain Res. 1983, 273, 133–141. [Google Scholar] [CrossRef]
- Steinfels, G.F.; Heym, J.; Strecker, R.E.; Jacobs, B.L. Behavioral correlates of dopaminergic unit activity in freely moving cats. Brain Res. 1983, 258, 217–228. [Google Scholar] [CrossRef]
- Xu, Y.L.; Reinscheid, R.K.; Huitron-Resendiz, S.; Clark, S.D.; Wang, Z.; Lin, S.H.; Brucher, F.A.; Zeng, J.; Ly, N.K.; Henriksen, S.J.; et al. Neuropeptide s: A neuropeptide promoting arousal and anxiolytic-like effects. Neuron 2004, 43, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Duangdao, D.M.; Clark, S.D.; Okamura, N.; Reinscheid, R.K. Behavioral phenotyping of neuropeptide s receptor knockout mice. Behav. Brain Res. 2009, 205, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jungling, K.; Seidenbecher, T.; Sosulina, L.; Lesting, J.; Sangha, S.; Clark, S.D.; Okamura, N.; Duangdao, D.M.; Xu, Y.L.; Reinscheid, R.K.; et al. Neuropeptide S-mediated control of fear expression and extinction: Role of intercalated GABAergic neurons in the amygdala. Neuron 2008, 59, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S. Physiological review article: Brain structures and mechanisms involved in the control of cortical activation and wakefulness, with emphasis on the posterior hypothalamus and histaminergic neurons. Sleep Med. Rev. 2000, 4, 471–503. [Google Scholar] [CrossRef] [PubMed]
- Denoyer, M.; Sallanon, M.; Buda, C.; Kitahama, K.; Jouvet, M. Neurotoxic lesion of the mesencephalic reticular formation and/or the posterior hypothalamus does not alter waking in the cat. Brain Res. 1991, 539, 287–303. [Google Scholar] [CrossRef]
- Gerashchenko, D.; Chou, T.C.; Blanco-Centurion, C.A.; Saper, C.B.; Shiromani, P.J. Effects of lesions of the histaminergic tuberomammillary nucleus on spontaneous sleep in rats. Sleep 2004, 27, 1275–1281. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.L.; Mochizuki, T.; Qu, W.M.; Hong, Z.Y.; Watanabe, T.; Urade, Y.; Hayaishi, O. Altered sleep–wake characteristics and lack of arousal response to h3 receptor antagonist in histamine h1 receptor knockout mice. Proc. Natl. Acad. Sci. USA 2006, 103, 4687–4692. [Google Scholar] [CrossRef] [PubMed]
- Parmentier, R.; Ohtsu, H.; Djebbara-Hannas, Z.; Valatx, J.L.; Watanabe, T.; Lin, J.S. Anatomical, physiological, and pharmacological characteristics of histidine decarboxylase knock-out mice: Evidence for the role of brain histamine in behavioral and sleep–wake control. J. Neurosci. 2002, 22, 7695–7711. [Google Scholar] [PubMed]
- Anaclet, C.; Parmentier, R.; Ouk, K.; Guidon, G.; Buda, C.; Sastre, J.P.; Akaoka, H.; Sergeeva, O.A.; Yanagisawa, M.; Ohtsu, H.; et al. Orexin/hypocretin and histamine: Distinct roles in the control of wakefulness demonstrated using knock-out mouse models. J. Neurosci. 2009, 29, 14423–14438. [Google Scholar] [CrossRef] [PubMed]
- Nishino, S.; Ripley, B.; Overeem, S.; Lammers, G.J.; Mignot, E. Hypocretin (orexin) deficiency in human narcolepsy. Lancet 2000, 355, 39–40. [Google Scholar] [CrossRef]
- Lin, L.; Faraco, J.; Li, R.; Kadotani, H.; Rogers, W.; Lin, X.Y.; Qiu, X.H.; de Jong, P.J.; Nishino, S.; Mignot, E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 1999, 98, 365–376. [Google Scholar] [CrossRef]
- Chemelli, R.M.; Willie, J.T.; Sinton, C.M.; Elmquist, J.K.; Scammell, T.; Lee, C.; Richardson, J.A.; Williams, S.C.; Xiong, Y.; Kisanuki, Y.; et al. Narcolepsy in orexin knockout mice: Molecular genetics of sleep regulation. Cell 1999, 98, 437–451. [Google Scholar] [CrossRef]
- Hara, J.; Beuckmann, C.T.; Nambu, T.; Willie, J.T.; Chemelli, R.M.; Sinton, C.M.; Sugiyama, F.; Yagami, K.; Goto, K.; Yanagisawa, M.; et al. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron 2001, 30, 345–354. [Google Scholar] [CrossRef]
- Piper, D.C.; Upton, N.; Smith, M.I.; Hunter, A.J. The novel brain neuropeptide, Orexin-A, modulates the sleep-wake cycle of rats. Eur. J. Neurosci. 2000, 12, 726–730. [Google Scholar] [CrossRef] [PubMed]
- Brisbare-Roch, C.; Dingemanse, J.; Koberstein, R.; Hoever, P.; Aissaoui, H.; Flores, S.; Mueller, C.; Nayler, O.; van Gerven, J.; de Haas, S.L.; et al. Promotion of sleep by targeting the orexin system in rats, dogs and humans. Nat. Med. 2007, 13, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Suzuki, M.; Mieda, M.; Tsujino, N.; Roth, B.; Sakurai, T. Pharmacogenetic modulation of orexin neurons alters sleep/wakefulness states in mice. PLoS ONE 2011, 6, e20360. [Google Scholar] [CrossRef] [PubMed]
- Munn, R.G.K.; Tyree, S.M.; McNaughton, N.; Bilkey, D.K. The frequency of hippocampal θ rhythm is modulated on a circadian period and is entrained by food availability. Front. Behav. Neurosci. 2015, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, M.; Yuasa, T.; Hayasaka, N.; Horikawa, K.; Sakurai, T.; Shibata, S. Reduced food anticipatory activity in genetically orexin (hypocretin) neuron-ablated mice. Eur. J. Neurosci. 2004, 20, 3054–3062. [Google Scholar] [CrossRef] [PubMed]
- Diano, S.; Horvath, B.; Urbanski, H.F.; Sotonyi, P.; Horvath, T.L. Fasting activates the nonhuman primate hypocretin (orexin) system and its postsynaptic targets. Endocrinology 2003, 144, 3774–3778. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, A.; Beuckmann, C.T.; Willie, J.T.; Hara, J.; Tsujino, N.; Mieda, M.; Tominaga, M.; Yagami, K.; Sugiyama, F.; Goto, K.; et al. Hypothalamic orexin neurons regulate arousal according to energy balance in mice. Neuron 2003, 38, 701–713. [Google Scholar] [CrossRef]
- Mieda, M.; Williams, S.C.; Sinton, C.M.; Richardson, J.A.; Sakurai, T.; Yanagisawa, M. Orexin neurons function in an efferent pathway of a food-entrainable circadian oscillator in eliciting food-anticipatory activity and wakefulness. J. Neurosci. 2004, 24, 10493–10501. [Google Scholar] [CrossRef] [PubMed]
- Rao, Y.; Lu, M.; Ge, F.; Marsh, D.J.; Qian, S.; Wang, A.H.; Picciotto, M.R.; Gao, X.B. Regulation of synaptic efficacy in hypocretin/orexin-containing neurons by melanin concentrating hormone in the lateral hypothalamus. J. Neurosci. 2008, 28, 9101–9110. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.Y.; Acuna-Goycolea, C.; van den Pol, A.N. Neuropeptide y inhibits hypocretin/orexin neurons by multiple presynaptic and postsynaptic mechanisms: Tonic depression of the hypothalamic arousal system. J. Neurosci. 2004, 24, 8741–8751. [Google Scholar] [CrossRef] [PubMed]
- Acuna-Goycolea, C.; van den Pol, A. Glucagon-like peptide 1 excites hypocretin/orexin neurons by direct and indirect mechanisms: Implications for viscera-mediated arousal. J. Neurosci. 2004, 24, 8141–8152. [Google Scholar] [CrossRef] [PubMed]
- Burdakov, D.; Jensen, L.T.; Alexopoulos, H.; Williams, R.H.; Fearon, I.M.; O’Kelly, I.; Gerasimenko, O.; Fugger, L.; Verkhratsky, A. Tandem-pore K+ channels mediate inhibition of orexin neurons by glucose. Neuron 2006, 50, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Ahnaou, A.; Drinkenburg, W.H.I.M.; Bouwknecht, J.A.; Alcazar, J.; Steckler, T.; Dautzenberg, F.M. Blocking melanin-concentrating hormone mch 1 receptor affects rat sleep–wake architecture. Eur. J. Pharmacol. 2008, 579, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Adamantidis, A.R.; Salvert, D.; Goutagny, R.; Lakaye, B.; Gervasoni, D.; Grisar, T.; Luppi, P.H.; Fort, P. Sleep architecture of the melanin–concentrating hormone receptor 1-knockout mice. Eur. J. Neurosci. 2008, 27, 1793–1800. [Google Scholar] [CrossRef] [PubMed]
- Willie, J.T.; Sinton, C.M.; Maratos-Flier, E.; Yanagisawa, M. Abnormal response of melanin-concentrating hormone deficient mice to fasting: Hyperactivity and rapid eye movement sleep suppression. Neuroscience 2008, 156, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Burdakov, D.; Gerasimenko, O.; Verkhratsky, A. Physiological changes in glucose differentially modulate the excitability of hypothalamic melanin-concentrating hormone and orexin neurons in situ. J. Neurosci. 2005, 25, 2429–2433. [Google Scholar] [CrossRef] [PubMed]
- Hur, E.E.; Zaborszky, L. Vglut2 afferents to the medial prefrontal and primary somatosensory cortices: A combined retrograde tracing in situ hybridization study [corrected]. J. Comp. Neurol. 2005, 483, 351–373. [Google Scholar] [CrossRef] [PubMed]
- Henny, P.; Jones, B.E. Projections from basal forebrain to prefrontal cortex comprise cholinergic, GABAergic and glutamatergic inputs to pyramidal cells or interneurons. Eur. J. Neurosci. 2008, 27, 654–670. [Google Scholar] [CrossRef] [PubMed]
- Gritti, I.; Mainville, L.; Mancia, M.; Jones, B.E. GABAergic and other noncholinergic basal forebrain neurons, together with cholinergic neurons, project to the mesocortex and isocortex in the rat. J. Comp. Neurol. 1997, 383, 163–177. [Google Scholar] [CrossRef]
- Freund, T.F.; Meskenaite, V. Γ-aminobutyric acid-containing basal forebrain neurons innervate inhibitory interneurons in the neocortex. Proc. Natl. Acad. Sci. USA 1992, 89, 738–742. [Google Scholar] [CrossRef] [PubMed]
- Bäckberg, M.; Ultenius, C.; Fritschy, J.M.; Meister, B. Cellular localization of GABA receptor α subunit immunoreactivity in the rat hypothalamus: Relationship with neurones containing orexigenic or anorexigenic peptides. J. Neuroendocrinol. 2004, 16, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Vincent, S.R.; Hokfelt, T.; Skirboll, L.R.; Wu, J.Y. Hypothalamic γ-aminobutyric acid neurons project to the neocortex. Science 1983, 220, 1309–1311. [Google Scholar] [CrossRef] [PubMed]
- Steffensen, S.C.; Svingos, A.L.; Pickel, V.M.; Henriksen, S.J. Electrophysiological characterization of GABAergic neurons in the ventral tegmental area. J. Neurosci. 1998, 18, 8003–8015. [Google Scholar] [PubMed]
- Rosin, D.L.; Weston, M.C.; Sevigny, C.P.; Stornetta, R.L.; Guyenet, P.G. Hypothalamic orexin (hypocretin) neurons express vesicular glutamate transporters vglut1 or vglut2. J. Comp. Neurol. 2003, 465, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.L.; Gall, C.M.; Jackson, V.R.; Civelli, O.; Reinscheid, R.K. Distribution of neuropeptide S receptor mRNA and neurochemical characteristics of neuropeptide S-expressing neurons in the rat brain. J. Comp. Neurol. 2007, 500, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Gras, C.; Herzog, E.; Bellenchi, G.C.; Bernard, V.; Ravassard, P.; Pohl, M.; Gasnier, B.; Giros, B.; El Mestikawy, S. A third vesicular glutamate transporter expressed by cholinergic and serotoninergic neurons. J. Neurosci. 2002, 22, 5442–5451. [Google Scholar] [PubMed]
- Kaur, S.; Pedersen, N.P.; Yokota, S.; Hur, E.E.; Fuller, P.M.; Lazarus, M.; Chamberlin, N.L.; Saper, C.B. Glutamatergic signaling from the parabrachial nucleus plays a critical role in hypercapnic arousal. J. Neurosci. 2013, 33, 7627–7640. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.N.; Kumar, S.; Suntsova, N.; Bashir, T.; Szymusiak, R.; McGinty, D. GABAergic regulation of the perifornical-lateral hypothalamic neurons during non-rapid eye movement sleep in rats. Neuroscience 2010, 167, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.S.; Steffensen, S.C.; Henriksen, S.J. Discharge profiles of ventral tegmental area GABA neurons during movement, anesthesia, and the sleep–wake cycle. J. Neurosci. 2001, 21, 1757–1766. [Google Scholar] [PubMed]
- Lu, J.; Greco, M.A.; Shiromani, P.; Saper, C.B. Effect of lesions of the ventrolateral preoptic nucleus on NREM and rem sleep. J. Neurosci. 2000, 20, 3830–3842. [Google Scholar] [PubMed]
- Szymusiak, R.; McGinty, D. Hypothalamic regulation of sleep and arousal. Ann. N. Y. Acad. Sci. 2008, 1129, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Adamantidis, A.R.; Zhang, F.; Aravanis, A.M.; Deisseroth, K.; de Lecea, L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 2007, 450, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; Adamantidis, A.; Ohtsu, H.; Deisseroth, K.; de Lecea, L. Sleep homeostasis modulates hypocretin-mediated sleep-wake transitions. J. Neurosci. 2009, 29, 10939–10949. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, T.; Kilduff, T.S.; Boyden, E.S.; Takahashi, S.; Tominaga, M.; Yamanaka, A. Acute optogenetic silencing of orexin/hypocretin neurons induces slow–wave sleep in mice. J. Neurosci. 2011, 31, 10529–10539. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, T.; Tabuchi, S.; Tanaka, K.F.; Boyden, E.S.; Tominaga, M.; Yamanaka, A. Long-lasting silencing of orexin/hypocretin neurons using archaerhodopsin induces slow-wave sleep in mice. Behav. Brain Res. 2013, 255, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Konadhode, R.R.; Pelluru, D.; Blanco-Centurion, C.; Zayachkivsky, A.; Liu, M.; Uhde, T.; Glen, W.B., Jr.; van den Pol, A.N.; Mulholland, P.J.; Shiromani, P.J. Optogenetic stimulation of mch neurons increases sleep. J. Neurosci. 2013, 33, 10257–10263. [Google Scholar] [CrossRef] [PubMed]
- Jego, S.; Glasgow, S.D.; Herrera, C.G.; Ekstrand, M.; Reed, S.J.; Boyce, R.; Friedman, J.; Burdakov, D.; Adamantidis, A.R. Optogenetic identification of a rapid eye movement sleep modulatory circuit in the hypothalamus. Nat. Neurosci. 2013, 16, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, T.; Ueno, T.; Tabuchi, S.; Inutsuka, A.; Tanaka, K.F.; Hasuwa, H.; Kilduff, T.S.; Terao, A.; Yamanaka, A. Optogenetic manipulation of activity and temporally controlled cell-specific ablation reveal a role for mch neurons in sleep/wake regulation. J. Neurosci. 2014, 34, 6896–6909. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.R.; Treweek, J.B.; Robinson, J.E.; Xiao, C.; Bremner, L.R.; Greenbaum, A.; Gradinaru, V. Dorsal raphe dopamine neurons modulate arousal and promote wakefulness by salient stimuli. Neuron 2017, 94, 1205–1219. [Google Scholar] [CrossRef] [PubMed]
- Van Dort, C.J.; Zachs, D.P.; Kenny, J.D.; Zheng, S.; Goldblum, R.R.; Gelwan, N.A.; Ramos, D.M.; Nolan, M.A.; Wang, K.; Weng, F.J. Optogenetic activation of cholinergic neurons in the PPT or LDT induces REM sleep. Proc. Natl. Acad. Sci. USA 2015, 112, 584–589. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.; Soya, S.; Sakurai, T. Excitation of GABAergic neurons in the bed nucleus of the stria terminalis triggers immediate transition from non-rapid eye movement sleep to wakefulness in mice. J. Neurosci. 2017, 37, 7164–7176. [Google Scholar] [CrossRef] [PubMed]
- Venner, A.; Anaclet, C.; Broadhurst, R.Y.; Saper, C.B.; Fuller, P.M. A novel population of wake-promoting GABAergic neurons in the ventral lateral hypothalamus. Curr. Biol. 2016, 26, 2137–2143. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Weber, F.; Zhong, P.; Tan, C.L.; Nguyen, T.N.; Beier, K.T.; Hörmann, N.; Chang, W.C.; Zhang, Z.; Do, J.P. Identification of preoptic sleep neurons using retrograde labelling and gene profiling. Nature 2017, 545, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Pelluru, D.; Konadhode, R.R.; Bhat, N.R.; Shiromani, P.J. Optogenetic stimulation of astrocytes in the posterior hypothalamus increases sleep at night in C57BL/6J mice. Eur. J. Neurosci. 2016, 43, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, K.S.; Sergeeva, O.; Brown, R.E.; Haas, H.L. Orexin/hypocretin excites the histaminergic neurons of the tuberomammillary nucleus. J Neurosci 2001, 21, 9273–9279. [Google Scholar] [PubMed]
- Nishino, S.; Sakurai, E.; Nevsimalova, S.; Yoshida, Y.; Watanabe, T.; Yanai, K.; Mignot, E. Decreased csf histamine in narcolepsy with and without low csf hypocretin-1 in comparison to healthy controls. Sleep 2009, 32, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Kanbayashi, T.; Kodama, T.; Kondo, H.; Satoh, S.; Inoue, Y.; Chiba, S.; Shimizu, T.; Nishino, S. Csf histamine contents in narcolepsy, idiopathic hypersomnia and obstructive sleep apnea syndrome. Sleep 2009, 32, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Nishino, S.; Fujiki, N.; Ripley, B.; Sakurai, E.; Kato, M.; Watanabe, T.; Mignot, E.; Yanai, K. Decreased brain histamine content in hypocretin/orexin receptor-2 mutated narcoleptic dogs. Neurosci. Lett. 2001, 313, 125–128. [Google Scholar] [CrossRef]
- Shigemoto, Y.; Fujii, Y.; Shinomiya, K.; Kamei, C. Participation of histaminergic H1 and noradrenergic α 1 receptors in orexin A-induced wakefulness in rats. Brain Res. 2004, 1023, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, A.; Tsujino, N.; Funahashi, H.; Honda, K.; Guan, J.L.; Wang, Q.P.; Tominaga, M.; Goto, K.; Shioda, S.; Sakurai, T. Orexins activate histaminergic neurons via the orexin 2 receptor. Biochem. Biophys. Res. Commun. 2002, 290, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.L.; Qu, W.M.; Li, W.D.; Mochizuki, T.; Eguchi, N.; Watanabe, T.; Urade, Y.; Hayaishi, O. Arousal effect of orexin a depends on activation of the histaminergic system. Proc. Natl. Acad. Sci. USA 2001, 98, 9965–9970. [Google Scholar] [CrossRef] [PubMed]
- Schone, C.; Cao, Z.F.; Apergis-Schoute, J.; Adamantidis, A.; Sakurai, T.; Burdakov, D. Optogenetic probing of fast glutamatergic transmission from hypocretin/orexin to histamine neurons in situ. J. Neurosci. 2012, 32, 12437–12443. [Google Scholar] [CrossRef] [PubMed]
- Schone, C.; Apergis-Schoute, J.; Sakurai, T.; Adamantidis, A.; Burdakov, D. Coreleased orexin and glutamate evoke nonredundant spike outputs and computations in histamine neurons. Cell Rep. 2014, 7, 697–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, R.H.; Chee, M.J.; Kroeger, D.; Ferrari, L.L.; Maratos-Flier, E.; Scammell, T.E.; Arrigoni, E. Optogenetic-mediated release of histamine reveals distal and autoregulatory mechanisms for controlling arousal. J. Neurosci. 2014, 34, 6023–6029. [Google Scholar] [CrossRef] [PubMed]
- Peyron, C.; Tighe, D.K.; van den Pol, A.N.; de Lecea, L.; Heller, H.C.; Sutcliffe, J.G.; Kilduff, T.S. Neurons containing hypocretin (orexin) project to multiple neuronal systems. J. Neurosci. 1998, 18, 9996–10015. [Google Scholar] [PubMed]
- Hagan, J.J.; Leslie, R.A.; Patel, S.; Evans, M.L.; Wattam, T.A.; Holmes, S.; Benham, C.D.; Taylor, S.G.; Routledge, C.; Hemmati, P.; et al. Orexin A activates locus coeruleus cell firing and increases arousal in the rat. Proc. Natl. Acad. Sci. USA 1999, 96, 10911–10916. [Google Scholar] [CrossRef] [PubMed]
- Bourgin, P.; Huitron–Resendiz, S.; Spier, A.D.; Fabre, V.; Morte, B.; Criado, J.R.; Sutcliffe, J.G.; Henriksen, S.J.; de Lecea, L. Hypocretin-1 modulates rapid eye movement sleep through activation of locus coeruleus neurons. J. Neurosci. 2000, 20, 7760–7765. [Google Scholar] [PubMed]
- Hasegawa, E.; Yanagisawa, M.; Sakurai, T.; Mieda, M. Orexin neurons suppress narcolepsy via 2 distinct efferent pathways. J. Clin. Investig. 2014, 124, 604–616. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; Yizhar, O.; Chikahisa, S.; Nguyen, H.; Adamantidis, A.; Nishino, S.; Deisseroth, K.; de Lecea, L. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 2010, 13, 1526–1533. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; Brill, J.; Bonnavion, P.; Huguenard, J.R.; Huerta, R.; de Lecea, L. Mechanism for hypocretin–mediated sleep-wake transitions. Proc. Natl. Acad. Sci. USA 2012, 109, E2635–E2644. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; de Lecea, L.; Adamantidis, A. Functional wiring of hypocretin and LC-NE neurons: Implications for arousal. Front. Behav. Neurosci. 2013, 7, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnavion, P.; Jackson, A.C.; Carter, M.E.; de Lecea, L. Antagonistic interplay between hypocretin and leptin in the lateral hypothalamus regulates stress responses. Nat. Commun. 2015, 6, 6266. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Garza, J.C.; Bronner, J.; Kim, C.S.; Zhang, W.; Lu, X.-Y. Acute administration of leptin produces anxiolytic-like effects: A comparison with fluoxetine. Psychopharmacology 2010, 207, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Tyree, S.M.; Munn, R.G.K.; McNaughton, N. Anxiolytic-like effects of leptin on fixed interval responding. Pharmacol. Biochem. Behav. 2016, 148, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Louis, G.W.; Leinninger, G.M.; Rhodes, C.J.; Myers, M.G. Direct innervation and modulation of orexin neurons by lateral hypothalamic LepRb neurons. J. Neurosci. 2010, 30, 11278–11287. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.; Tritos, N.A.; Lowell, B.B.; Flier, J.S.; Maratos–Flier, E. Mice lacking melanin-concentrating hormone are hypophagic and lean. Nature 1998, 396, 670–674. [Google Scholar] [PubMed]
- Marsh, D.J.; Weingarth, D.T.; Novi, D.E.; Chen, H.Y.; Trumbauer, M.E.; Chen, A.S.; Guan, X.M.; Jiang, M.M.; Feng, Y.; Camacho, R.E. Melanin-concentrating hormone 1 receptor-deficient mice are lean, hyperactive, and hyperphagic and have altered metabolism. Proc. Natl. Acad. Sci. USA 2002, 99, 3240–3245. [Google Scholar] [CrossRef] [PubMed]
- Takase, K.; Kikuchi, K.; Tsuneoka, Y.; Oda, S.; Kuroda, M.; Funato, H. Meta-analysis of melanin–concentrating hormone signaling-deficient mice on behavioral and metabolic phenotypes. PLoS ONE 2014, 9, e99961. [Google Scholar] [CrossRef] [PubMed]
- Apergis-Schoute, J.; Iordanidou, P.; Faure, C.; Jego, S.; Schone, C.; Aitta-Aho, T.; Adamantidis, A.; Burdakov, D. Optogenetic evidence for inhibitory signaling from orexin to mch neurons via local microcircuits. J. Neurosci. 2015, 35, 5435–5441. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Zhou, J.; Feng, Q.; Lin, R.; Gong, H.; Luo, Q.; Zeng, S.; Luo, M.; Fu, L. Multi–channel fiber photometry for population neuronal activity recording. Biomed. Opt. Express 2015, 6, 3919–3931. [Google Scholar] [CrossRef] [PubMed]
- Adelsberger, H.; Grienberger, C.; Stroh, A.; Konnerth, A. In vivo calcium recordings and channelrhodopsin-2 activation through an optical fiber. Cold Spring Harb. Protoc. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Basu, A.; Satija, R.; Nemesh, J.; Shekhar, K.; Goldman, M.; Tirosh, I.; Bialas, A.R.; Kamitaki, N.; Martersteck, E.M. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015, 161, 1202–1214. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.N.; Macosko, E.Z.; Fenselau, H.; Pers, T.H.; Lyubetskaya, A.; Tenen, D.; Goldman, M.; Verstegen, A.M.J.; Resch, J.M.; McCarroll, S.A. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 2017, 20, 484–496. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tyree, S.M.; De Lecea, L. Optogenetic Investigation of Arousal Circuits. Int. J. Mol. Sci. 2017, 18, 1773. https://doi.org/10.3390/ijms18081773
Tyree SM, De Lecea L. Optogenetic Investigation of Arousal Circuits. International Journal of Molecular Sciences. 2017; 18(8):1773. https://doi.org/10.3390/ijms18081773
Chicago/Turabian StyleTyree, Susan M., and Luis De Lecea. 2017. "Optogenetic Investigation of Arousal Circuits" International Journal of Molecular Sciences 18, no. 8: 1773. https://doi.org/10.3390/ijms18081773




