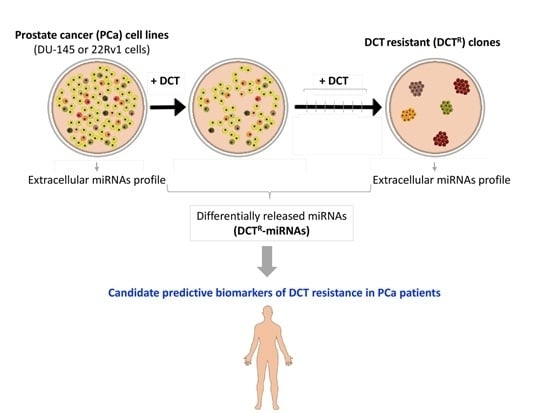
The Prostate Cancer Cells Resistant to Docetaxel as in vitro Model for Discovering MicroRNAs Predictive of the Onset of Docetaxel Resistance
Abstract
:1. Introduction
2. Results
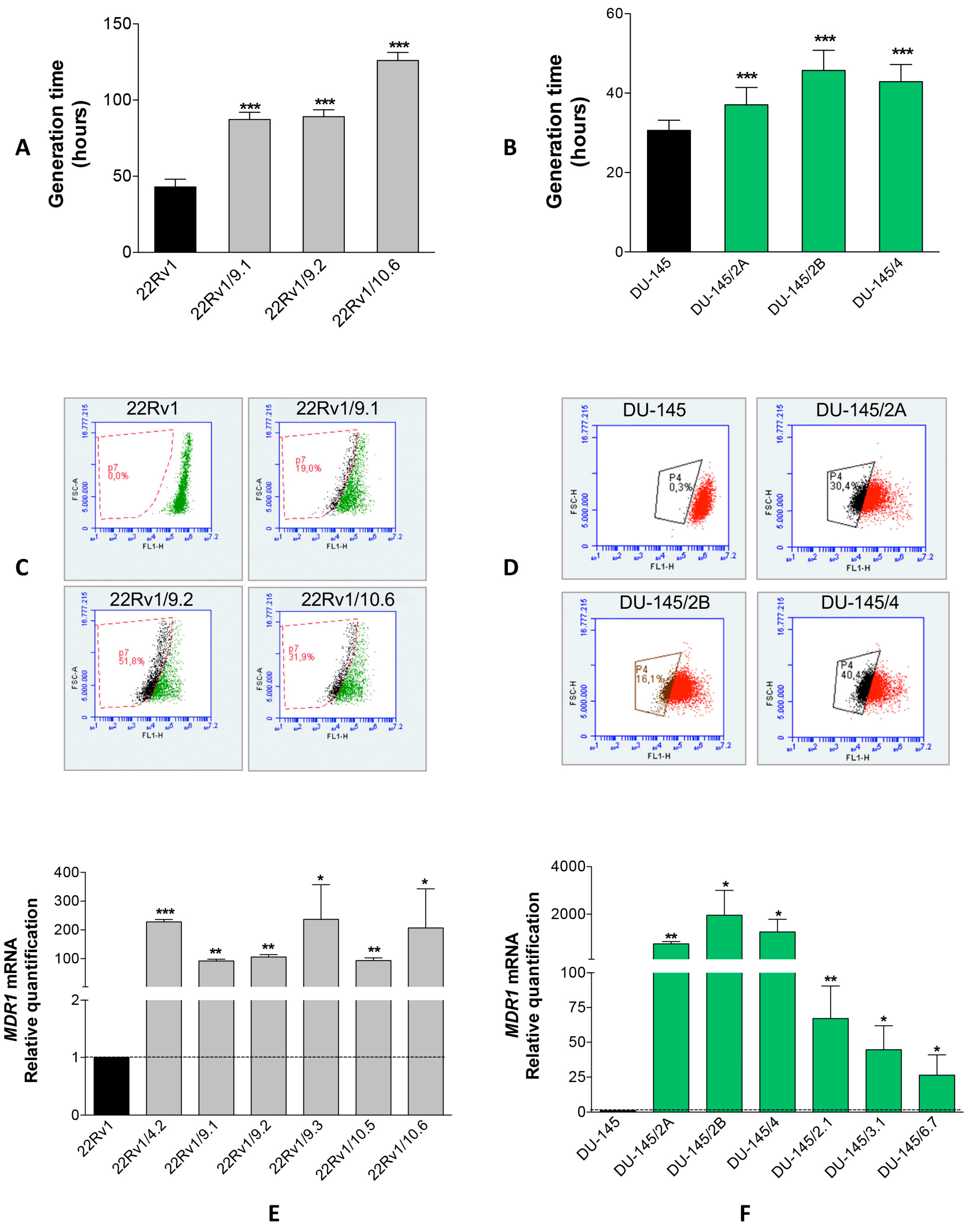
2.1. Isolation of DCTR Clones from 22Rv1 and DU-145 Prostate Cancer Cell Lines
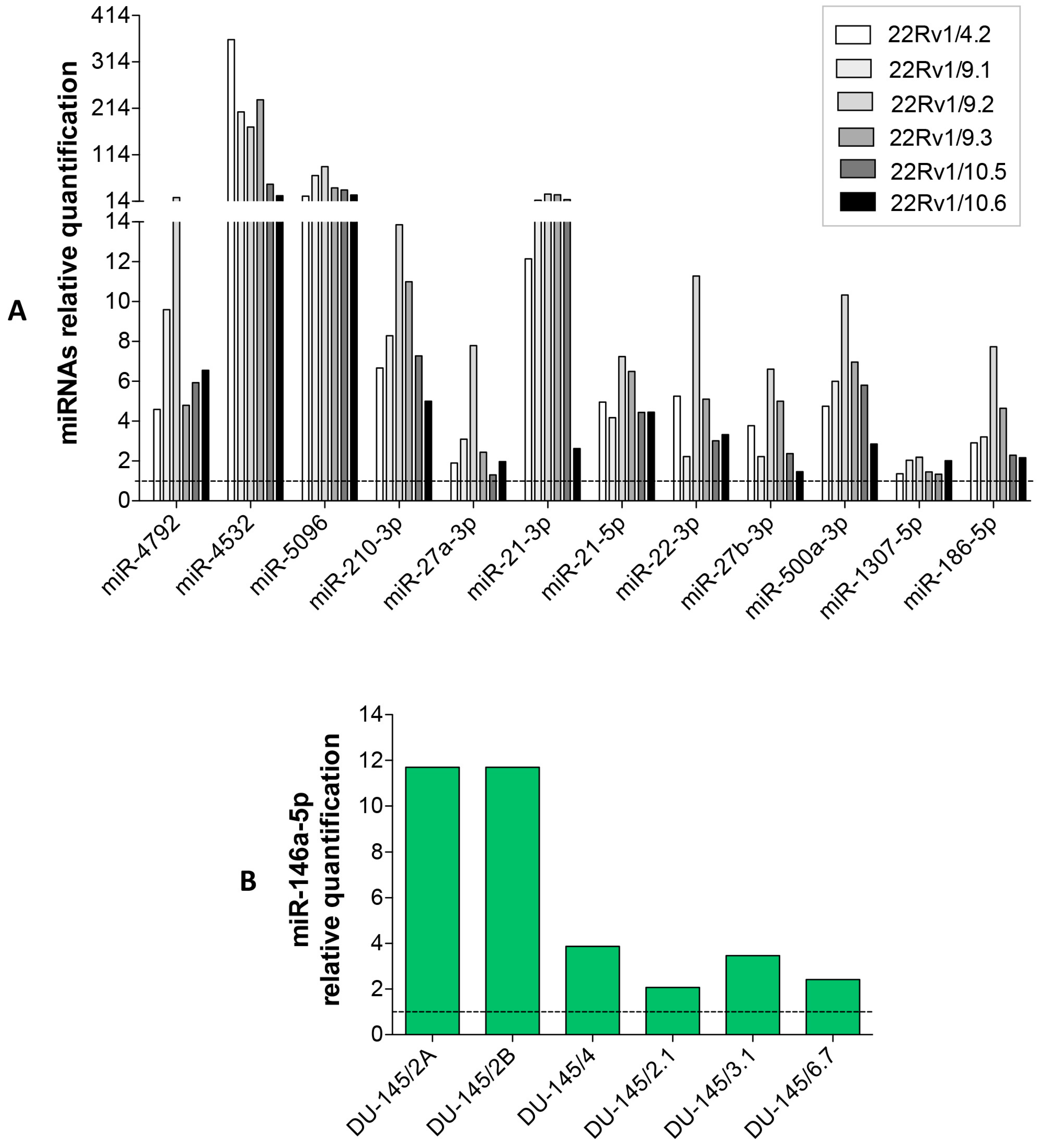
2.2. Identification of miRNAs Differentially Released by DCTR Clones
2.3. DCTR-miRNAs as Possible Biomarkers of DCT Resistance
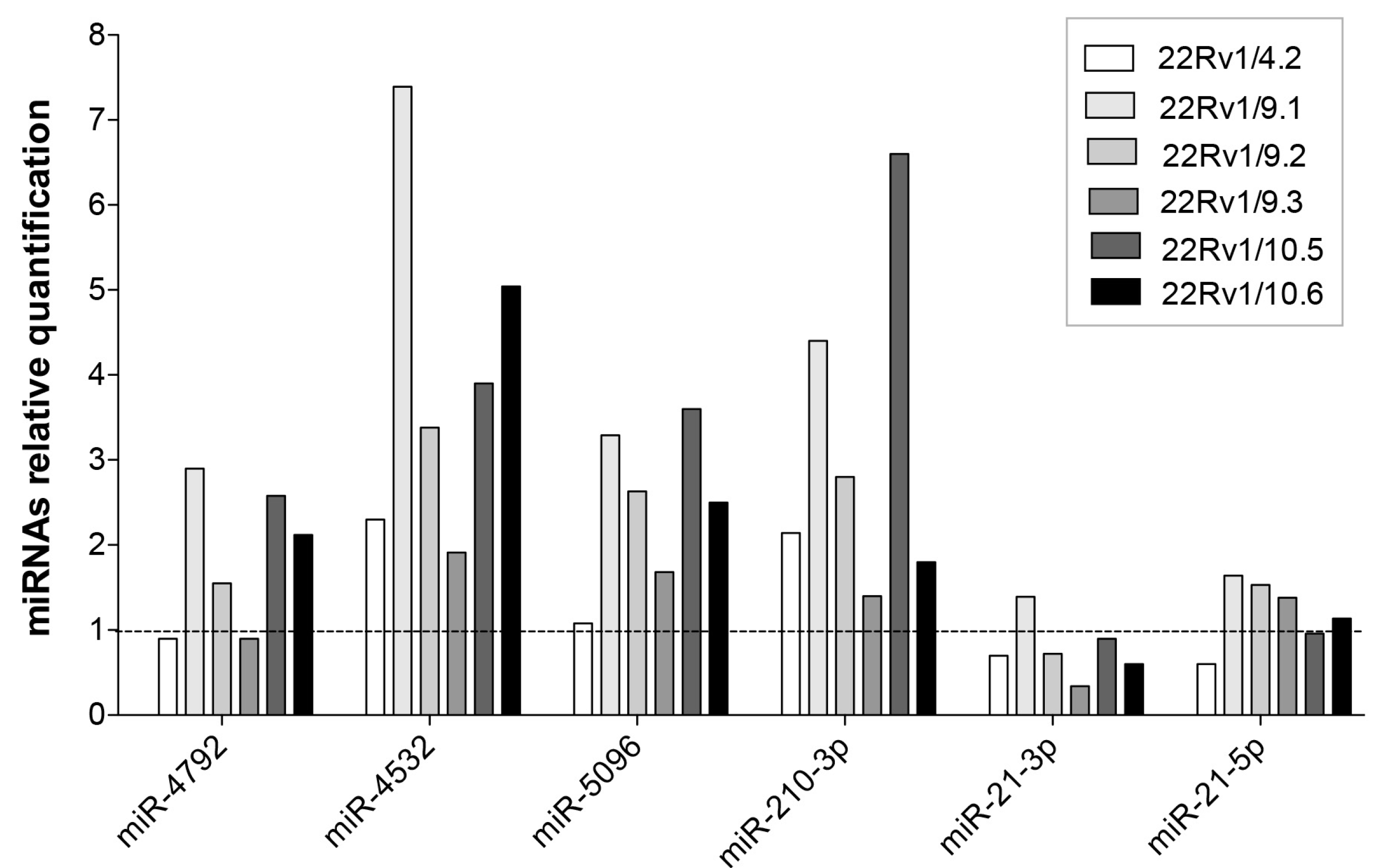
2.4. Expression Level of Selected DCTR-miRNAs in DCTR Clones
3. Discussion
4. Materials and Methods
4.1. Cells and Culture Conditions
4.2. Dose Response Curves
4.3. Isolation and Characterization of DCT-Resistant (DCTR) Clones
4.4. Rhodamine 123 Exclusion Assay
4.5. Total RNA Isolation
4.6. miRNAs and mRNA Quantification
4.7. miRNA Profiling with Next-Generation Sequencing (NGS) Technology (Small RNA-Seq)
4.8. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schurko, B.; Oh, W.K. Docetaxel chemotherapy remains the standard of care in castration-resistant prostate cancer. Nat. Clin. Pract. Oncol. 2008, 5, 506–507. [Google Scholar] [CrossRef] [PubMed]
- Petrylak, D.P.; Tangen, C.M.; Hussain, M.H.; Lara, P.N., Jr.; Jones, J.A.; Taplin, M.E.; Burch, P.A.; Berry, D.; Moinpour, C.; Kohli, M.; et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N. Engl. J. Med. 2004, 351, 1513–1520. [Google Scholar] [CrossRef] [PubMed]
- Endzelins, E.; Melne, V.; Kalnina, Z.; Lietuvietis, V.; Riekstina, U.; Llorente, A.; Line, A. Diagnostic, prognostic and predictive value of cell-free miRNAs in prostate cancer: A systematic review. Mol. Cancer 2016, 15, 41. [Google Scholar] [CrossRef] [PubMed]
- Filella, X.; Foj, L. miRNAs as novel biomarkers in the management of prostate cancer. Clin. Chem. Lab. Med. 2017, 55, 715–736. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.P.; Shen, Y.J.; Shi, G.H.; Ye, D.W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Mitchell, P.S.; Kroh, E.M.; Dowell, A.E.; Chery, L.; Siddiqui, J.; Nelson, P.S.; Vessella, R.L.; Knudsen, B.S.; Chinnaiyan, A.M.; et al. Circulating microRNA profiling identifies a subset of metastatic prostate cancer patients with evidence of cancer-associated hypoxia. PLoS ONE 2013, 8, e69239. [Google Scholar] [CrossRef] [PubMed]
- Lucotti, S.; Rainaldi, G.; Evangelista, M.; Rizzo, M. Fludarabine treatment favors the retention of miR-485-3p by prostate cancer cells: Implications for survival. Mol. Cancer 2013, 12, 52. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, K.; Rocchi, P.; Miyake, H.; Fazli, L.; So, A.; Zangemeister-Wittke, U.; Gleave, M.E. Induction of apoptosis and enhancement of chemosensitivity in human prostate cancer LNCaP cells using bispecific antisense oligonucleotide targeting Bcl-2 and Bcl-xL genes. BJU Int. 2006, 97, 1300–1308. [Google Scholar] [CrossRef] [PubMed]
- Terry, S.; Ploussard, G.; Allory, Y.; Nicolaiew, N.; Boissiere-Michot, F.; Maille, P.; Kheuang, L.; Coppolani, E.; Ali, A.; Bibeau, F.; et al. Increased expression of class III beta-tubulin in castration-resistant human prostate cancer. Br. J. Cancer 2009, 101, 951–956. [Google Scholar] [CrossRef] [PubMed]
- Sartor, O.; Gillessen, S. Treatment sequencing in metastatic castrate-resistant prostate cancer. Asian J. Androl. 2014, 16, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Hathaway, A.R.; Baker, M.K.; Sonpavde, G. Emerging agents for the therapy of advanced prostate cancer. Future Oncol. 2015, 11, 2775–2787. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G. Circulating miRNAs: Roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. 2015, 81, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef] [PubMed]
- Armand-Labit, V.; Pradines, A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol. Concepts 2017, 8, 61–81. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, J.C.; Fink, L.M.; Goodman, O.B., Jr.; Symanowski, J.T.; Vogelzang, N.J.; Ward, D.C. Comparison of circulating MicroRNA 141 to circulating tumor cells, lactate dehydrogenase, and prostate-specific antigen for determining treatment response in patients with metastatic prostate cancer. Clin. Genitourin. Cancer 2011, 9, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Townley, S.L.; Bert, A.G.; Stricker, P.D.; Sutherland, P.D.; Horvath, L.G.; Goodall, G.J.; Butler, L.M.; Tilley, W.D. Circulating microRNAs predict biochemical recurrence in prostate cancer patients. Br. J. Cancer 2013, 109, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Castillo, L.; Mahon, K.L.; Chiam, K.; Lee, B.Y.; Nguyen, Q.; Boyer, M.J.; Stockler, M.R.; Pavlakis, N.; Marx, G.; et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br. J. Cancer 2014, 110, 2462–2471. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Mahon, K.L.; Spielman, C.; Gurney, H.; Mallesara, G.; Stockler, M.R.; Bastick, P.; Briscoe, K.; Marx, G.; Swarbrick, A.; et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br. J. Cancer 2017, 116, 1002–1011. [Google Scholar] [CrossRef] [PubMed]
- James, N.D.; Sydes, M.R.; Clarke, N.W.; Mason, M.D.; Dearnaley, D.P.; Spears, M.R.; Ritchie, A.W.; Parker, C.C.; Russell, J.M.; Attard, G.; et al. Addition of docetaxel, zoledronic acid, or both to first-line long-term hormone therapy in prostate cancer (STAMPEDE): Survival results from an adaptive, multiarm, multistage, platform randomised controlled trial. Lancet 2016, 387, 1163–1177. [Google Scholar] [CrossRef]
- O’Neill, A.J.; Prencipe, M.; Dowling, C.; Fan, Y.; Mulrane, L.; Gallagher, W.M.; O’Connor, D.; O’Connor, R.; Devery, A.; Corcoran, C.; et al. Characterisation and manipulation of docetaxel resistant prostate cancer cell lines. Mol. Cancer 2011, 10, 126. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, C.; Nadiminty, N.; Lou, W.; Tummala, R.; Evans, C.P.; Gao, A.C. Inhibition of ABCB1 expression overcomes acquired docetaxel resistance in prostate cancer. Mol. Cancer Ther. 2013, 12, 1829–1836. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef] [PubMed]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and extracellular microRNA: An update on localization and biological role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Tonevitsky, A.G.; Burwinkel, B. Extracellular miRNA: A collision of two paradigms. Trends Biochem. Sci. 2016, 41, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Le, Q.T.; Giaccia, A.J. MiR-210-micromanager of the hypoxia pathway. Trends Mol. Med. 2010, 16, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Bertout, J.A.; Patel, S.A.; Simon, M.C. The impact of O2 availability on human cancer. Nat. Rev. Cancer 2008, 8, 967–975. [Google Scholar] [CrossRef] [PubMed]
- Taddei, M.L.; Cavallini, L.; Comito, G.; Giannoni, E.; Folini, M.; Marini, A.; Gandellini, P.; Morandi, A.; Pintus, G.; Raspollini, M.R.; et al. Senescent stroma promotes prostate cancer progression: The role of miR-210. Mol. Oncol. 2014, 8, 1729–1746. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Sin, W.C.; Harris, A.L.; Naus, C.C. Gap junctions modulate glioma invasion by direct transfer of microRNA. Oncotarget 2015, 6, 15566. [Google Scholar] [CrossRef] [PubMed]
- Thuringer, D.; Boucher, J.; Jego, G.; Pernet, N.; Cronier, L.; Hammann, A.; Solary, E.; Garrido, C. Transfer of functional microRNAs between glioblastoma and microvascular endothelial cells through gap junctions. Oncotarget 2016, 7, 73925–73934. [Google Scholar] [CrossRef] [PubMed]
- Touil, Y.; Zuliani, T.; Wolowczuk, I.; Kuranda, K.; Prochazkova, J.; Andrieux, J.; Le Roy, H.; Mortier, L.; Vandomme, J.; Jouy, N.; et al. The PI3K/AKT signaling pathway controls the quiescence of the low-Rhodamine123-retention cell compartment enriched for melanoma stem cell activity. Stem Cells 2013, 31, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Barsanti, C.; Trivella, M.G.; D’Aurizio, R.; El Baroudi, M.; Baumgart, M.; Groth, M.; Caruso, R.; Verde, A.; Botta, L.; Cozzi, L.; et al. Differential regulation of microRNAs in end-stage failing hearts is associated with left ventricular assist device unloading. BioMed Res. Int. 2015, 2015, 592512. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wang, W.C.; Lin, F.M.; Chang, W.C.; Lin, K.Y.; Huang, H.D.; Lin, N.S. miRExpress: Analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinform. 2009, 10, 328. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]



| DCTR/miRNAs | Column A *. DCTR-miRNAs Level in DCTR Clones Versus Parental Cells Growth Medium | Column B **. DCTR-miRNAs Level in DCT-Treated Versus Untreated Cells Growth Medium | Column A/Column B | Unpaired t-Test Pvalue (A Versus B) |
|---|---|---|---|---|
| miR-4792 *** | 8.94 ± 2.75 | 1.69 ± 0.11 | 5.27 | p < 0.05 |
| miR-4532 | 175.20 ± 50.66 | 2.10 ± 0.48 | 83.42 | p < 0.01 |
| miR-5096 | 48.61 ± 10.00 | 1.28 ± 0.03 | 37.97 | p < 0.001 |
| miR-210-3p | 8.80 ± 1.13 | 2.28 ± 0.19 | 3.86 | p < 0.001 |
| miR-27a-3p | 3.48 ± 0.85 | 2.13 ± 0.2 | 1.63 | NS |
| miR-21-3p | 17.7 ± 4.12 | 3.9 ± 0.49 | 4.53 | p < 0.01 |
| miR-21-5p | 5.18 ± 0.43 | 2.49 ± 0.39 | 2.07 | p < 0.001 |
| miR-22-3p | 5.22 ± 1.21 | 3.86 ± 0.32 | 1.35 | NS |
| miR-27b-3p | 3.52 ± 0.77 | 4.29 ± 0.25 | 0.82 | NS |
| miR-500a-3p | 6.11 ± 1.01 | 3.78 ± 0.03 | 1.61 | p < 0.05 |
| miR-186-5p | 3.82 ± 0.86 | 3.64 ± 0.43 | 1.05 | NS |
| miR-146a-5p **** | 5.87 ± 1.86 | 10.21 ± 1.46 | 0.57 | NS |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bascetta, L.; Oliviero, A.; D’Aurizio, R.; Evangelista, M.; Mercatanti, A.; Pellegrini, M.; Marrocolo, F.; Bracarda, S.; Rizzo, M. The Prostate Cancer Cells Resistant to Docetaxel as in vitro Model for Discovering MicroRNAs Predictive of the Onset of Docetaxel Resistance. Int. J. Mol. Sci. 2017, 18, 1512. https://doi.org/10.3390/ijms18071512
Bascetta L, Oliviero A, D’Aurizio R, Evangelista M, Mercatanti A, Pellegrini M, Marrocolo F, Bracarda S, Rizzo M. The Prostate Cancer Cells Resistant to Docetaxel as in vitro Model for Discovering MicroRNAs Predictive of the Onset of Docetaxel Resistance. International Journal of Molecular Sciences. 2017; 18(7):1512. https://doi.org/10.3390/ijms18071512
Chicago/Turabian StyleBascetta, Lorenzo, Arianna Oliviero, Romina D’Aurizio, Monica Evangelista, Alberto Mercatanti, Marco Pellegrini, Francesca Marrocolo, Sergio Bracarda, and Milena Rizzo. 2017. "The Prostate Cancer Cells Resistant to Docetaxel as in vitro Model for Discovering MicroRNAs Predictive of the Onset of Docetaxel Resistance" International Journal of Molecular Sciences 18, no. 7: 1512. https://doi.org/10.3390/ijms18071512