Multi-Approach Analysis for the Identification of Proteases within Birch Pollen
Abstract
:1. Introduction
2. Results
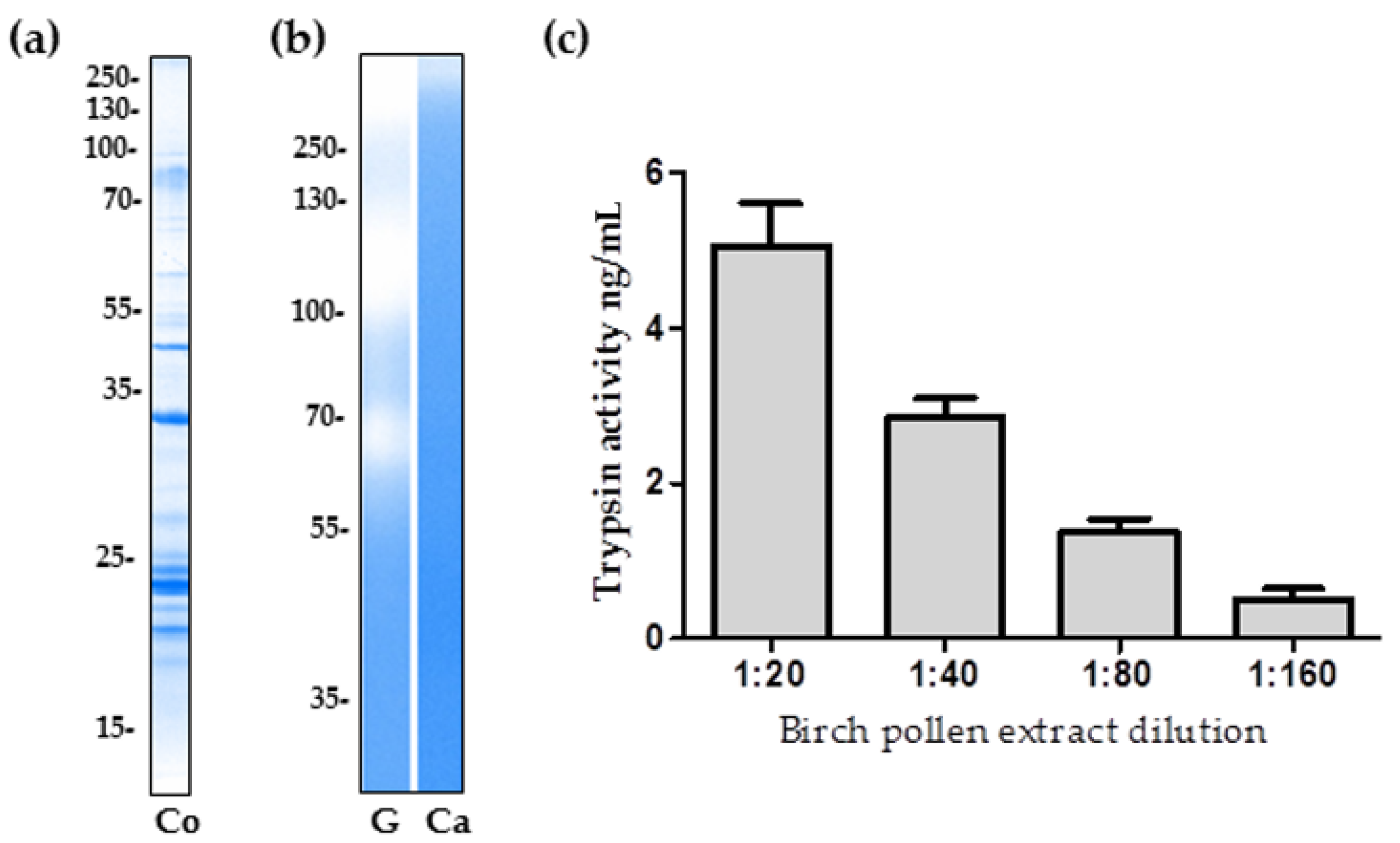
2.1. The Proteolytic Activity of Birch Pollen Extract
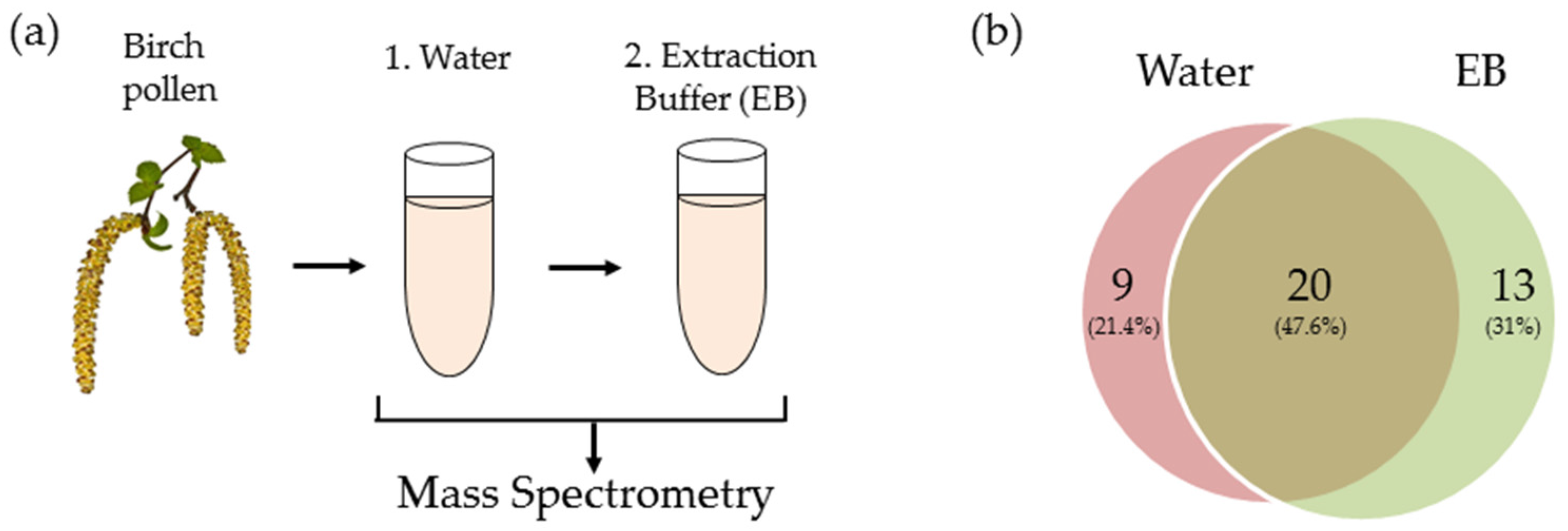
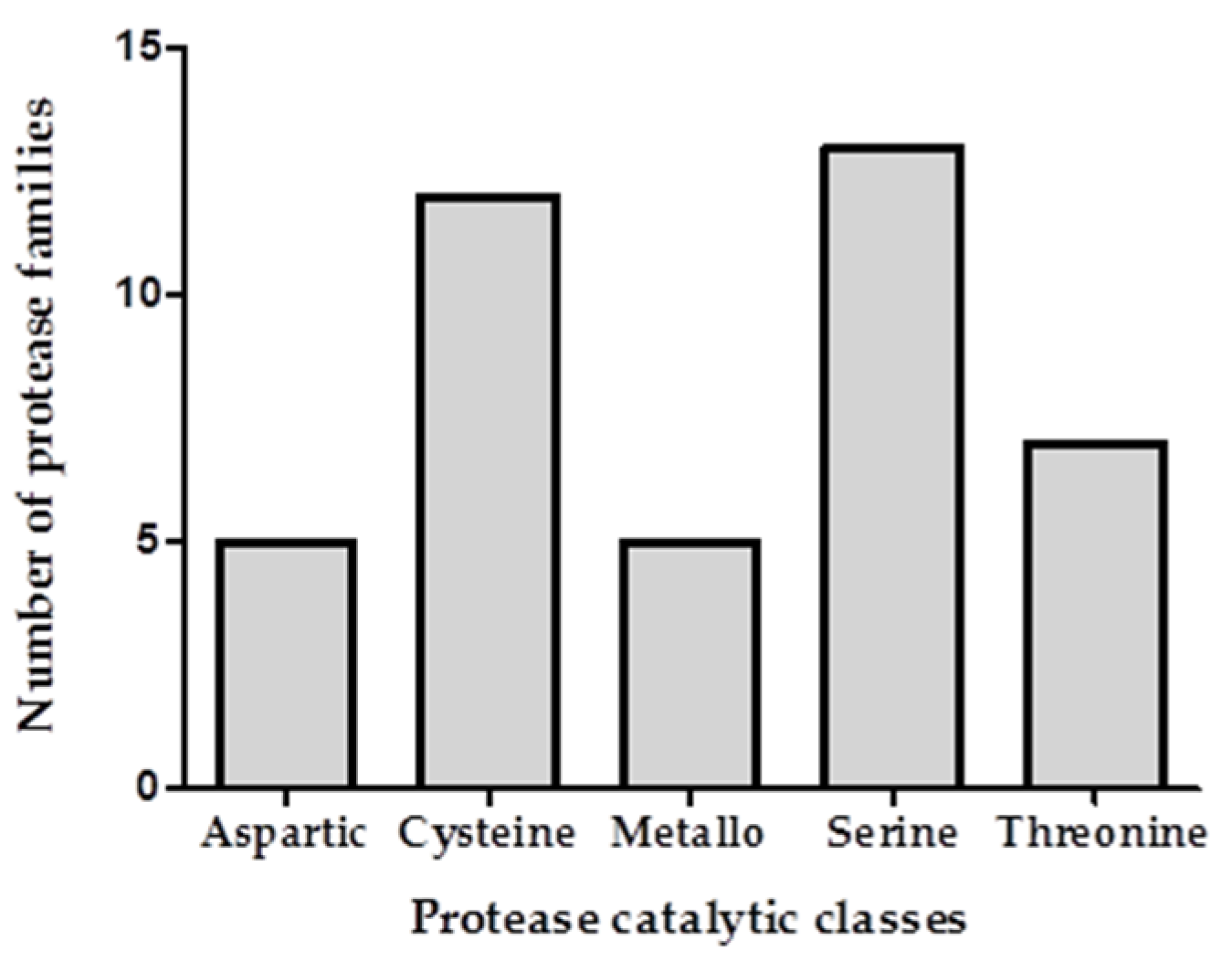
2.2. Transcriptomic and Proteomic Analysis
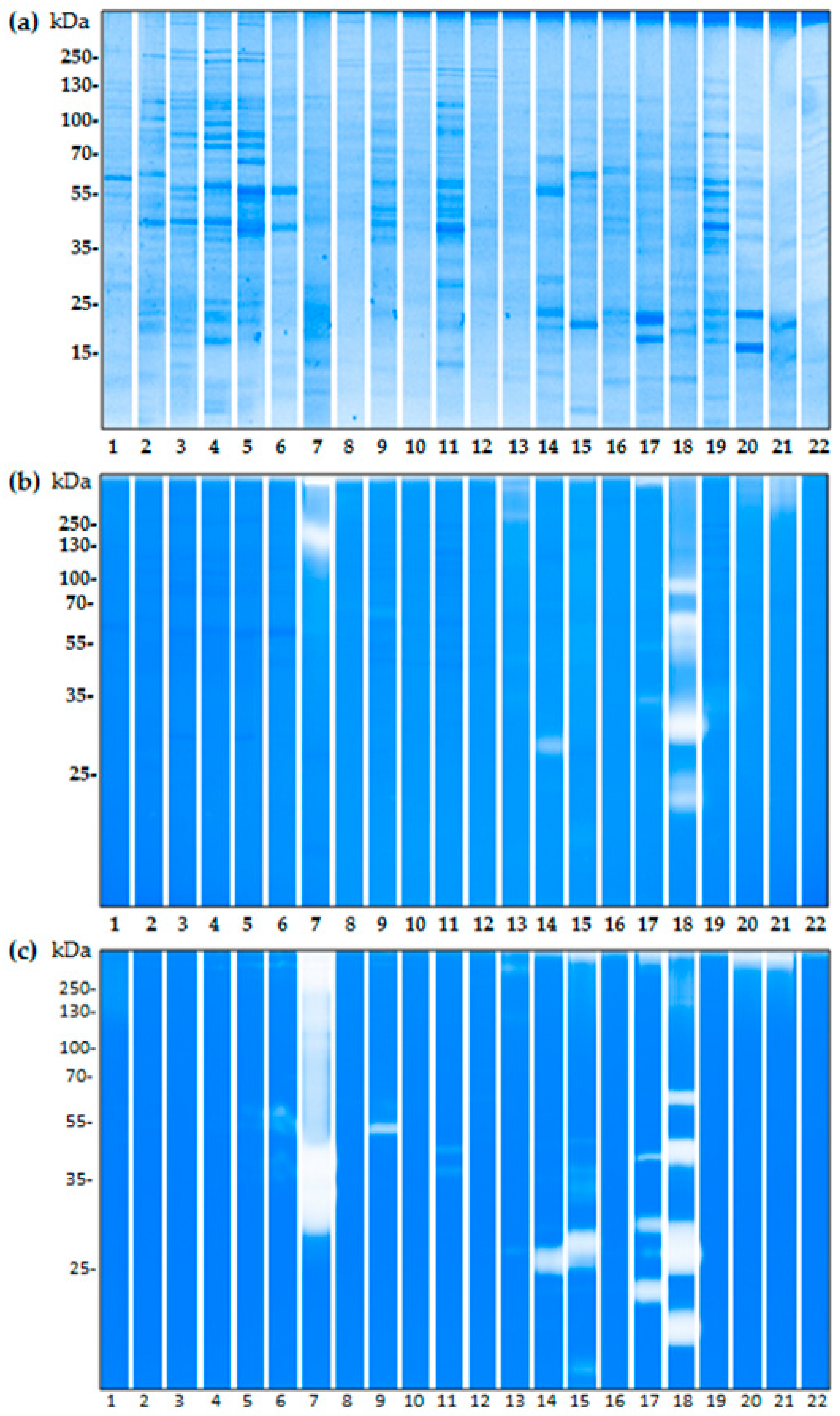
2.3. The Proteolytic Activity of Bacterial Isolates from Birch Pollen
3. Discussion
4. Materials and Methods
4.1. Pollen Extract Preparation
4.2. Isolation of Bacteria on Pollen Grains
4.3. SDS-PAGE
4.4. Zymography
4.5. Protease Activity Assay
4.6. RNA Sequencing
4.7. Mass Spectrometry Analysis
4.8. Further Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van der Hoorn, R.A.L. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.B.; Tanksale, A.M.; Ghatge, M.S.; Deshpande, V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998, 62, 597–635. [Google Scholar] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the merops database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44, D343–D350. [Google Scholar] [CrossRef] [PubMed]
- Krichevskya, A.; Kozlovsky, S.V.; Tian, G.W.; Chen, M.H.; Zaltsmana, A.; Citovskya, V. How pollen tubes grow. Dev. Biol. 2007, 303, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Widmer, F.; Hayes, P.J.; Whittaker, R.G.; Kumar, R.K. Substrate preference profiles of proteases released by allergenic pollens. Clin. Exp. Allergy 2000, 30, 571–576. [Google Scholar] [CrossRef] [PubMed]
- Blanchard, G.C.; Gardner, R. Characterization of some of enzymes in ragweed pollen. Ann. Allergy 1976, 36, 410–418. [Google Scholar] [PubMed]
- Hassim, Z.; Maronese, S.E.; Kumar, R.K. Injury to murine airway epithelial cells by pollen enzymes. Thorax 1998, 53, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Baraniuk, J.N.; Esch, R.E.; Buckley, C.E. Pollen grain column chromatography—Quantitation and biochemical-analysis of ragweed-pollen solutes. J. Allergy Clin. Immun. 1988, 81, 1126–1134. [Google Scholar] [CrossRef]
- Bouley, J.; Groeme, R.; le Mignon, M.; Jain, K.; Chabre, H.; Bordas-Le Floch, V.; Couret, M.N.; Bussieres, L.; Lautrette, A.; Naveau, M.; et al. Identification of the cysteine protease amb a 11 as a novel major allergen from short ragweed. J. Allergy Clin. Immun. 2015, 136, 1055–1064. [Google Scholar] [CrossRef] [PubMed]
- Vinhas, R.; Cortes, L.; Cardoso, I.; Mendes, V.M.; Manadas, B.; Todo-Bom, A.; Pires, E.; Verissimo, P. Pollen proteases compromise the airway epithelial barrier through degradation of transmembrane adhesion proteins and lung bioactive peptides. Allergy 2011, 66, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y. Role of allergen source-derived proteases in sensitization via airway epithelial cells. J. Allergy 2012, 2012, 903659. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, K.; McAllister, H.A. Botanical Magazine Monograph. In The Genus Betula: A Taxonomic Revision of Birches; Illustrated Reprint; Royal Botanic Gardens, Kew publishing: Kew, UK, 2013; Volume 5, p. 432. [Google Scholar]
- Hollbacher, B.; Schmitt, A.O.; Hofer, H.; Ferreira, F.; Lackner, P. Identification of proteases and protease inhibitors in allergenic and non-allergenic pollen. Int. J. Mol. Sci. 2017, 18, 1199. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.E.; Kita, H. The role of protease activation of inflammation in allergic respiratory diseases. J. Allergy Clin. Immun. 2004, 114, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.A.; Holgate, S.T. The airway epithelium: Structural and functional properties in health and disease. Respirology 2003, 8, 432–446. [Google Scholar] [CrossRef] [PubMed]
- Runswick, S.; Mitchell, T.; Davies, P.; Robinson, C.; Garrod, D.R. Pollen proteolytic enzymes degrade tight junctions. Respirology 2007, 12, 834–842. [Google Scholar] [CrossRef] [PubMed]
- Enjoji, S.; Ohama, T.; Sato, K. Regulation of epithelial cell tight junctions by protease-activated receptor 2. J. Vet. Med. Sci. 2014, 76, 1225–1229. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zeng, X.; He, S. Evaluation on potential contributions of protease activated receptors related mediators in allergic inflammation. Mediators Inflamm. 2014, 2014, 829068. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Bateman, A. Merops: The peptidase database. Nucleic Acids Res. 2010, 38, D227–D233. [Google Scholar] [CrossRef] [PubMed]
- Kamphuis, I.G.; Kalk, K.H.; Swarte, M.B.A.; Drenth, J. Structure of papain refined at 1.65 Å resolution. J. Mol. Biol. 1984, 179, 233–256. [Google Scholar] [CrossRef]
- Vannella, K.M.; Ramalingam, T.R.; Hart, K.M.; Prado, R.D.; Sciurba, J.; Barron, L.; Borthwick, L.A.; Smith, A.D.; Mentink-Kane, M.; White, S.; et al. Acidic chitinase primes the protective immune response to gastrointestinal nematodes. Nat. Immunol. 2016, 17, 538. [Google Scholar] [CrossRef] [PubMed]
- Furmonaviciene, R.; Ghaemmaghami, A.M.; Boyd, S.E.; Jones, N.S.; Bailey, K.; Willis, A.C.; Sewell, H.F.; Mitchell, D.A.; Shakib, F. The protease allergen Der p 1 cleaves cell surface DC-SIGN and DC-SIGNR: Experimental analysis of in silico substrate identification and implications in allergic responses. Clin. Exp. Allergy 2007, 37, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.L.; Chen, X.C.; Liu, F.; Chen, W.; Wu, P.; Wieschhaus, A.J.; Chishti, A.H.; Roche, P.A.; Chen, W.M.; Lin, T.J. Calpain-1 contributes to ige-mediated mast cell activation. J. Immunol. 2014, 192, 5130–5139. [Google Scholar] [CrossRef] [PubMed]
- Litosh, V.A.; Rochman, M.; Rymer, J.K.; Porollo, A.; Kottyan, L.C.; Rothenberg, M.E. Calpain-14 and its association with eosinophilic esophagitis. J. Allergy Clin. Immunol. 2017, 139, 1762–1771. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and timps. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [PubMed]
- Ubl, J.J.; Grishina, Z.V.; Sukhomlin, T.K.; Welte, T.; Sedehizade, F.; Reiser, G. Human bronchial epithelial cells express PAR-2 with different sensitivity to thermolysin. Am. J. Physiol. Lung Cell. Mol. Physiol. 2002, 282, L1339–L1348. [Google Scholar] [CrossRef] [PubMed]
- Cera, E.D. Serine proteases. IUBMB Life 2009, 61, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, S.; Mizuguchi, Y.; Morimoto, K.; Aki, T.; Shigeta, S.; Yasueda, H.; Wada, T.; Suzuki, O.; Jyo, T.; Ono, K. Cloning and expression of Der f 6, a serine protease allergen from the house dust mite, dermatophagoides farinae. BBA-Mol. Basis Dis. 1999, 1454, 201–207. [Google Scholar] [CrossRef]
- Gupta, R.; Sharma, V.; Sridhara, S.; Singh, B.P.; Arora, N. Identification of serine protease as a major allergen of curvularia lunata. Allergy 2004, 59, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Bagarozzi, D.A.; Potempa, J.; Travis, J. Purification and characterization of an arginine-specific peptidase from ragweed (Ambrosia artemisiifolia) pollen. Am. J. Resp. Cell. Mol. 1998, 18, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Bagarozzi, D.A., Jr.; Pike, R.; Potempa, J.; Travis, J. Purification and characterization of a novel endopeptidase in ragweed (Ambrosia artemisiifolia) pollen. J. Biol. Chem. 1996, 271, 26227–26232. [Google Scholar] [CrossRef] [PubMed]
- Florsheim, E.; Yu, S.; Bragatto, I.; Faustino, L.; Gomes, E.; Ramos, R.N.; Barbuto, J.A.; Medzhitov, R.; Russo, M. Integrated innate mechanisms involved in airway allergic inflammation to the serine protease subtilisin. J. Immunol. 2015, 194, 4621–4630. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Nair, S.; Singh, B.P.; Arora, N. Molecular and immunological characterization of subtilisin like serine protease, a major allergen of curvularia lunata. Immunobiology 2011, 216, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Beg, Q.K.; Lorenz, P. Bacterial alkaline proteases: Molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002, 59, 15–32. [Google Scholar] [PubMed]
- Poll, V.; Denk, U.; Shen, H.D.; Panzani, R.C.; Dissertori, O.; Lackner, P.; Hemmer, W.; Mari, A.; Crameri, R.; Lottspeich, F.; et al. The vacuolar serine protease, a cross-reactive allergen from cladosporium herbarum. Mol. Immunol. 2009, 46, 1360–1373. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T.; Church, M.K.; Broide, D.H.; Martinez, F.D. Allergy, 4th ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Montealegre, F.; Fernandez, B.; Delgado, A.; Fernandez, L.; Roman, A.; Chardon, D.; Rodriguez-Santana, J.; Medina, V.; Zavala, D.; Bayona, M. Exposure levels of asthmatic children to allergens, endotoxins, and serine proteases in a tropical environment. J. Asthma 2004, 41, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Obersteiner, A.; Gilles, S.; Frank, U.; Beck, I.; Haring, F.; Ernst, D.; Rothballer, M.; Hartmann, A.; Traidl-Hoffmann, C.; Schmid, M. Pollen-associated microbiome correlates with pollution parameters and the allergenicity of pollen. PLoS ONE 2016, 11, e0149545. [Google Scholar] [CrossRef] [PubMed]
- Manirajan, B.A.; Ratering, S.; Rusch, V.; Schwiertz, A.; Geissler-Plaum, R.; Cardinale, M.; Schnell, S. Bacterial microbiota associated with flower pollen is influenced by pollination type, and shows a high degree of diversity and species-specificity. Environ. Microbiol. 2016, 18, 5161–5174. [Google Scholar] [CrossRef] [PubMed]
- Pepys, J.; Longbottom, J.L.; Hargreave, F.E.; Faux, J. Allergic reactions of the lungs to enzymes of Bacillus subtilis. Lancet 1969, 1, 1181–1184. [Google Scholar] [CrossRef]
- Dolovich, J.; Little, D.C. Correlates of skin-test reactions to Bacillus-subtilis enzyme preparations. J. Allergy Clin. Immun. 1972, 49, 43–53. [Google Scholar] [CrossRef]
- Basketter, D.A.; Kruszewski, F.H.; Mathieu, S.; Kirchner, D.B.; Panepinto, A.; Fieldsend, M.; Siegert, V.; Barnes, F.; Bookstaff, R.; Simonsen, M.; et al. Managing the risk of occupational allergy in the enzyme detergent industry. J. Occup. Environ. Hyg. 2015, 12, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Khatri, I.; Sharma, S.; Ramya, T.N.C.; Subramanian, S. Complete genomes of Bacillus coagulans S-lac and Bacillus subtilis TO-A JPC, two phylogenetically distinct probiotics. PLoS ONE 2016, 11, e0156745. [Google Scholar] [CrossRef] [PubMed]
- Prabha, M.S.; Divakar, K.; Priya, J.D.A.; Selvam, G.P.; Balasubramanian, N.; Gautam, P. Statistical analysis of production of protease and esterase by a newly isolated lysinibacillus fusiformis AU01: Purification and application of protease in sub-culturing cell lines. Ann. Microbiol. 2015, 65, 33–46. [Google Scholar] [CrossRef]
- Wandersman, C. Secretion, processing and activation of bacterial extracellular proteases. Mol. Microbiol. 1989, 3, 1825–1831. [Google Scholar] [CrossRef] [PubMed]
- Jackson, R.M.; Russell, R.B. Predicting function from structure: Examples of the serine protease inhibitor canonical loop conformation found in extracellular proteins. Comput. Chem. 2001, 26, 31–39. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-Length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
| Catalytic Type | Description | MEROPS Accession | Clan (Subclan) | Family (Subfamily) | Ext. |
|---|---|---|---|---|---|
| Aspartic | Aspartic proteinase A1 | A01.A02 | AA | A1 (A) | EB, W |
| Aspartic | Aspartic proteinase CDR1 | A01.069 | AA | A1 (B) | EB, W |
| Aspartic | Aspartic proteinase oryzasin-1 | A01.020 | AA | A1 (A) | EB |
| Aspartic | β-secretase 2 | A01.041 | AA | A1 (A) | W |
| Aspartic | Signal peptide peptidase | A22.A15 | AD | A22 (B) | W |
| Cysteine | Calpain-type cysteine protease ADL1 | C02.019 | CA | C2 (A) | EB |
| Cysteine | Cysteine protease RD19A | C01.022 | CA | C1 (A) | W |
| Cysteine | Cysteine proteinase RD21A | C01.064 | CA | C1 (A) | EB, W |
| Cysteine | Desumoylating isopeptidase 1 | C97.001 | CP | C97 | EB |
| Cysteine | OTU domain-containing protein 5 | C85.001 | CA | C85 (B) | EB |
| Cysteine | Protein DJ-1 homolog D | C56.A01 | PC (C) | C56 | EB, W |
| Cysteine | Thiol protease aleurain-like | C01.162 | CA | C19 (A) | W |
| Cysteine | Ubiquitin carboxyl-terminal hydrolase | C12.A03 | CA | C12 | EB, W |
| Cysteine | Ubiquitin carboxyl-terminal hydrolase | C19.094 | CA | C19 | EB, W |
| Cysteine | Ubiquitin carboxyl-terminal hydrolase | C19.068 | CA | C19 | W |
| Cysteine | Ubiquitin thioesterase OTU1 | C85.007 | CA | C85 (B) | EB |
| Cysteine | Ubiquitin-like-specific protease 1D | C48.A04 | CE | C48 | EB |
| Metallo | Leucine aminopeptidase 2 | n/a | MF | M17 | EB, W |
| Metallo | Mitochondrial-peptidase subunit α 2 | n/a | n/a | n/a | EB, W |
| Metallo | Organellar oligopeptidase A | M03.A01 | MA (E) | M3 (A) | EB, W |
| Metallo | Probable Xaa-Pro aminopeptidase P | M24.A10 | MG | M24 (B) | EB, W |
| Metallo | Puromycin-sensitive aminopeptidase | M01.029 | MA (E) | M1 | EB, W |
| Serine | ATP-dependent Clp protease subunit 5 | S14.A01 | SK | S14 | EB |
| Serine | Dipeptidyl peptidase family member 6 | S09.A77 | SC | S9 | EB, W |
| Serine | Probable glutamyl endopeptidase, | S09.021 | SC | S9 (D) | EB |
| Serine | Serine carboxypeptidase-like 20 | S10.A11 | SC | S10 | EB, W |
| Serine | Serine carboxypeptidase-like 40 | S10.A41 | SC | S10 | W |
| Serine | Serine carboxypeptidase-like 42 | S10.A21 | SC | S10 | W |
| Serine | Serine carboxypeptidase-like 48 | S10.A46 | SC | S10 | EB, W |
| Serine | Serine carboxypeptidase-like 49 | S10.A45 | SC | S10 | EB, W |
| Serine | Subtilisin-like protease SBT1.7 | S08.112 | SB | S8 (A) | EB, W |
| Serine | Subtilisin-like protease SBT1.8 | S08.A24 | SB | S8 (A) | EB |
| Serine | Subtilisin-like protease SBT4.15 | S08.A13 | SB | S8 (A) | EB |
| Serine | Subtilisin-like protease SBT5.4 | S08.A26 | SB | S8 (A) | EB, W |
| Serine | Tripeptidyl-peptidase 2 | S08.A56 | SB | S8 (A) | EB |
| Threonine | Proteasome subunit α type-3 | n/a | n/a | n/a | W |
| Threonine | Proteasome subunit α type-5-B | T01.995 | PB (T) | T1 (A) | EB, W |
| Threonine | Proteasome subunit α type-6 | T01.971 | PB (T) | T1 (A) | EB, W |
| Threonine | Proteasome subunit α type-6-B | n/a | n/a | n/a | EB |
| Threonine | Proteasome subunit α type-7 | n/a | PB (T) | T1 (A) | EB, W |
| Threonine | Proteasome subunit β type-4 | T01.987 | PB (T) | T1 (X) | W |
| Threonine | Proteasome subunit β type-5-B | T01.A10 | PB (T) | T1 (A) | EB |
| Gram Stain | No. in Gel | Bacterial Order | Bacterial Family | C | G |
|---|---|---|---|---|---|
| negative | 1 | Caulobacterales | Caulobacteraceae | – | + |
| 2 | Enterobacteriales | Noctuoideaceae | – | – | |
| 3 | Pseudomonadales | Pseudomonadaceae | – | – | |
| 4 | Pseudomonadales | Pseudomonadaceae | – | – | |
| 5 | Pseudomonadales | Pseudomonadaceae | – | – | |
| 6 | Sphingomonadales | Sphingomonadaceae | – | + | |
| 7 | Xanthomonadales | Xanthomonadaceae | ++ | +++ | |
| positive | 8 | Actinomycetales | Gordoniaceae | – | – |
| 9 | Actinomycetales | Microbacteriaceae | + | + | |
| 10 | Actinomycetales | Micrococcaceae | – | – | |
| 11 | Actinomycetales | Micrococcaceae | – | + | |
| 12 | Actinomycetales | Nocardioidaceae | – | – | |
| 13 | Actinomycetales | Streptomycetaceae | + | + | |
| 14 | Bacillales | Bacillaceae | + | ++ | |
| 15 | Bacillales | Bacillaceae | – | ++ | |
| 16 | Bacillales | Bacillaceae | – | – | |
| 17 | Bacillales | Bacillaceae | + | ++ | |
| 18 | Bacillales | Bacillaceae | ++ | +++ | |
| 19 | Bacillales | Bacillaceae | + | – | |
| 20 | Bacillales | Bacillaceae | – | + | |
| 21 | Bacillales | Bacillaceae | + | + | |
| 22 | Bacillales | Paenibacillaceae | – | – |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McKenna, O.E.; Posselt, G.; Briza, P.; Lackner, P.; Schmitt, A.O.; Gadermaier, G.; Wessler, S.; Ferreira, F. Multi-Approach Analysis for the Identification of Proteases within Birch Pollen. Int. J. Mol. Sci. 2017, 18, 1433. https://doi.org/10.3390/ijms18071433
McKenna OE, Posselt G, Briza P, Lackner P, Schmitt AO, Gadermaier G, Wessler S, Ferreira F. Multi-Approach Analysis for the Identification of Proteases within Birch Pollen. International Journal of Molecular Sciences. 2017; 18(7):1433. https://doi.org/10.3390/ijms18071433
Chicago/Turabian StyleMcKenna, Olivia E., Gernot Posselt, Peter Briza, Peter Lackner, Armin O. Schmitt, Gabriele Gadermaier, Silja Wessler, and Fatima Ferreira. 2017. "Multi-Approach Analysis for the Identification of Proteases within Birch Pollen" International Journal of Molecular Sciences 18, no. 7: 1433. https://doi.org/10.3390/ijms18071433







