Cellular Consequences of Diminished Protein O-Mannosyltransferase Activity in Baker’s Yeast
Abstract
:1. Introduction
2. Results
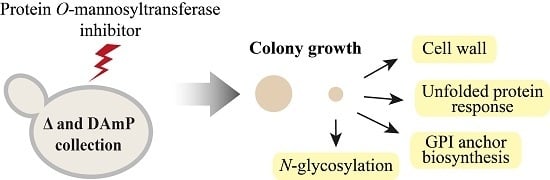
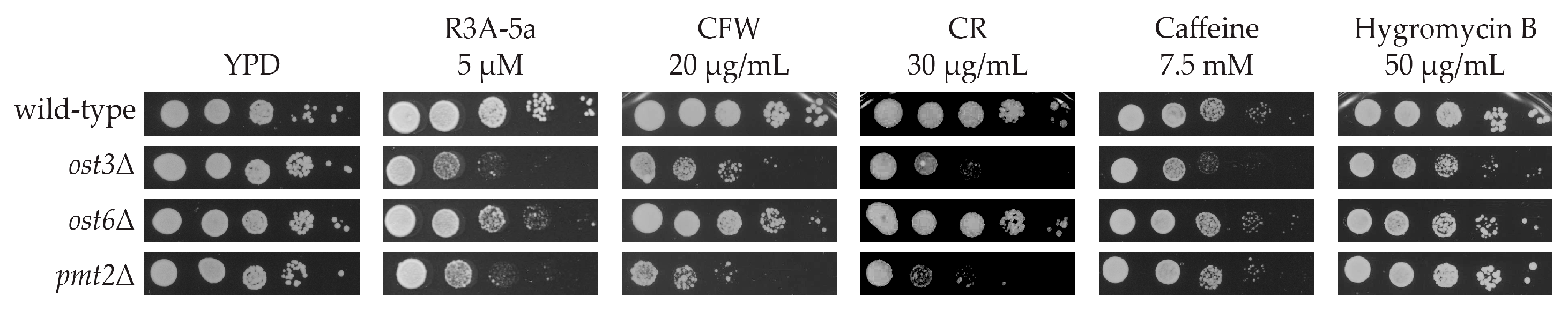
2.1. Systematic Analysis of Factors Important for Normal Cell Growth upon Impaired Protein O-Mannosylation
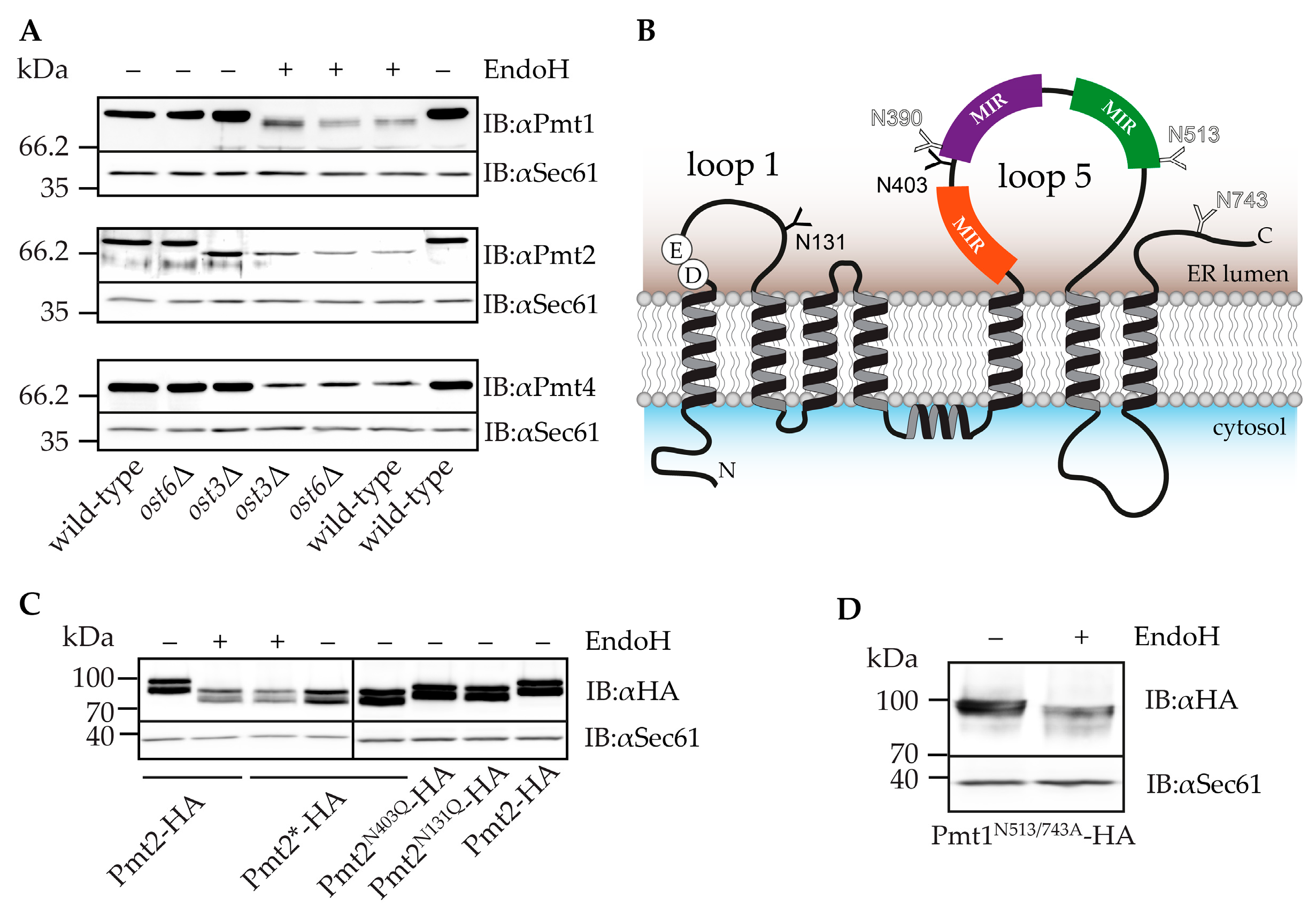
2.2. Pmt2 N-Glycans are Ost3 Specific
2.3. Impact of N-Glycosylation on Pmt1-Pmt2 Activity
3. Discussion
4. Materials and Methods
4.1. Yeast Strains and Culture Conditions
4.2. R3A-5a Screen
4.3. Spotting Assay
4.4. Preparation of Total Membranes and Endoglycosidase H Treatment
4.5. Isolation of Heat Shock Protein 150 and Chitinase
4.6. SDS-PAGE and Immunoblot
4.7. Flow Cytometry
4.8. In Vitro Mannosyltransferase Activity Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| ER | endoplasmic reticulum |
| Dol-P-Man | dolichol phosphate-activated mannose |
| PMT | protein O-mannosyltransferase |
| OST | oligosaccharyltransferase |
| GDP-mannose | guanosine diphosphate-activated mannose |
| CWIP | cell wall integrity pathway |
| GPI | glycophosphatidylinositol |
| GFP | green fluorescent protein |
| GO | gene ontology |
| EndoH | endoglycosidase H |
| HA | hemagglutinin |
References
- Gentzsch, M.; Tanner, W. The PMT gene family: Protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996, 15, 5752–5759. [Google Scholar] [PubMed]
- Willer, T.; Brandl, M.; Sipiczki, M.; Strahl, S. Protein O-mannosylation is crucial for cell wall integrity, septation and viability in fission yeast. Mol. Microbiol. 2005, 57, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Hennet, T.; Cabalzar, J. Congenital disorders of glycosylation: A concise chart of glycocalyx dysfunction. Trends Biochem Sci. 2015, 40, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Neubert, P.; Strahl, S. Protein O-mannosylation in the early secretory pathway. Curr. Opin. Cell Biol. 2016, 41, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Wunderle, L.; Hutzler, J.; Schulz, B.L.; Aebi, M.; Strahl, S. Protein O-mannosyltransferases associate with the translocon to modify translocating polypeptide chains. J. Biol. Chem. 2014, 289, 8599–8611. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Mrsa, V.; Hagen, I.; Deutzmann, R.; Strahl, S.; Tanner, W. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 2003, 4, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Neubert, P.; Halim, A.; Zauser, M.; Essig, A.; Joshi, H.J.; Zatorska, E.; Larsen, I.S.; Loibl, M.; Castells-Ballester, J.; Aebi, M.; et al. Mapping the O-Mannose Glycoproteome in Saccharomyces cerevisiae. Mol. Cell. Proteom. 2016, 15, 1323–1337. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Scheinost, A. Transmembrane topology of Pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 1999, 274, 9068–9075. [Google Scholar] [CrossRef]
- Girrbach, V.; Strahl, S. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 2003, 278, 12554–12562. [Google Scholar] [CrossRef] [PubMed]
- Lommel, M.; Schott, A.; Jank, T.; Hofmann, V.; Strahl, S. A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J. Biol. Chem. 2011, 286, 39768–39775. [Google Scholar] [CrossRef] [PubMed]
- Bausewein, D.; Engel, J.; Jank, T.; Schoedl, M.; Strahl, S. Functional Similarities between the Protein O-Mannosyltransferases Pmt4 from Bakers’ Yeast and Human POMT1. J. Biol. Chem. 2016, 291, 18006–18015. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Immervoll, T.; Deutzmann, R.; Tanner, W. PMT1, the gene for a key enzyme of protein O-glycosylation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1993, 90, 8164–8168. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.; Hutzler, J.; Bermejo, C.; Ragni, E.; Garcia-Cantalejo, J.; Botias, P.; Piberger, H.; Schott, A.; Sanz, A.B.; Strahl, S. Functional and genomic analyses of blocked protein O-mannosylation in baker’s yeast. Mol. Microbiol. 2011, 79, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Klis, F.M.; de Koster, C.G.; Brul, S. Cell wall-related bionumbers and bioestimates of Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 2014, 13, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Goder, V.; Melero, A. Protein O-mannosyltransferases participate in ER protein quality control. J. Cell Sci. 2011, 124, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ng, D.T. O-mannosylation: The other glycan player of ER quality control. Semi. Cell Dev. Biol. 2015, 41, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Wang, S.; Thibault, G.; Ng, D.T. Futile protein folding cycles in the ER are terminated by the unfolded protein O-mannosylation pathway. Science 2013, 340, 978–981. [Google Scholar] [CrossRef] [PubMed]
- Orchard, M.G.; Neuss, J.C.; Galley, C.M.; Carr, A.; Porter, D.W.; Smith, P.; Scopes, D.I.; Haydon, D.; Vousden, K.; Stubberfield, C.R.; et al. Rhodanine-3-acetic acid derivatives as inhibitors of fungal protein mannosyl transferase 1 (PMT1). Bioorg. Med. Chem. Lett. 2004, 14, 3975–3978. [Google Scholar] [CrossRef] [PubMed]
- Tabas-Madrid, D.; Nogales-Cadenas, R.; Pascual-Montano, A. GeneCodis3: A non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012, 40, W478–W483. [Google Scholar] [CrossRef] [PubMed]
- Nogales-Cadenas, R.; Carmona-Saez, P.; Vazquez, M.; Vicente, C.; Yang, X.; Tirado, F.; Carazo, J.M.; Pascual-Montano, A. GeneCodis: Interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009, 37, W317–W322. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Saez, P.; Chagoyen, M.; Tirado, F.; Carazo, J.M.; Pascual-Montano, A. GENECODIS: A web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. Evol. 2007, 8, R3. [Google Scholar] [CrossRef] [PubMed]
- Cherry, J.M.; Hong, E.L.; Amundsen, C.; Balakrishnan, R.; Binkley, G.; Chan, E.T.; Christie, K.R.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; et al. Saccharomyces Genome Database: The genomics resource of budding yeast. Nucleic Acids Res. 2012, 40, D700–D705. [Google Scholar] [CrossRef] [PubMed]
- Roumanie, O.; Peypouquet, M.F.; Bonneu, M.; Thoraval, D.; Doignon, F.; Crouzet, M. Evidence for the genetic interaction between the actin-binding protein Vrp1 and the RhoGAP Rgd1 mediated through Rho3p and Rho4p in Saccharomyces cerevisiae. Mol. Microbiol. 2000, 36, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Zatorska, E.; Strahl, S.; Centre for Organismal Studies, Heidelberg University, Heidelberg, Germany. Gene ontology cellular component and molecular function analysis using GeneCodis. 2017. [Google Scholar]
- Sanz, A.B.; Garcia, R.; Rodriguez-Pena, J.M.; Nombela, C.; Arroyo, J. Cooperation between SAGA and SWI/SNF complexes is required for efficient transcriptional responses regulated by the yeast MAPK Slt2. Nucleic Acids Res. 2016, 44, 7159–7172. [Google Scholar] [CrossRef] [PubMed]
- Sanz, A.B.; Garcia, R.; Rodriguez-Pena, J.M.; Diez-Muniz, S.; Nombela, C.; Peterson, C.L.; Arroyo, J. Chromatin remodeling by the SWI/SNF complex is essential for transcription mediated by the yeast cell wall integrity MAPK pathway. Mol. Biol. Cell 2012, 23, 2805–2817. [Google Scholar] [CrossRef] [PubMed]
- Thewes, S. Calcineurin-Crz1 signaling in lower eukaryotes. Eukaryot. Cell 2014, 13, 694–705. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.E.; Cyert, M.S. The eukaryotic response regulator Skn7p regulates calcineurin signaling through stabilization of Crz1p. EMBO J. 2001, 20, 3473–3483. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Irie, K.; Matsumoto, K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol. Cell. Biol. 1995, 15, 5740–5749. [Google Scholar] [CrossRef] [PubMed]
- Lavina, W.A.; Hermansyah; Sugiyama, M.; Kaneko, Y.; Harashima, S. Functionally redundant protein phosphatase genes PTP2 and MSG5 co-regulate the calcium signaling pathway in Saccharomyces cerevisiae upon exposure to high extracellular calcium concentration. J. Biosci. Bioeng. 2013, 115, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Mattison, C.P.; Spencer, S.S.; Kresge, K.A.; Lee, J.; Ota, I.M. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 1999, 19, 7651–7660. [Google Scholar] [CrossRef] [PubMed]
- Stanger, K.; Gorelik, M.; Davidson, A.R. Yeast adaptor protein, Nbp2p, is conserved regulator of fungal Ptc1p phosphatases and is involved in multiple signaling pathways. J. Biol. Chem. 2012, 287, 22133–22141. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Scrimale, T.; Didone, L.; de Mesy Bentley, K.L.; Krysan, D.J. The unfolded protein response is induced by the cell wall integrity mitogen-activated protein kinase signaling cascade and is required for cell wall integrity in Saccharomyces cerevisiae. Mol. Biol. Cell 2009, 20, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.L.; Aebi, M. Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol. Cell. Proteomics 2009, 8, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, K.; Leberre, V.; Sokol, S.; Palamarczyk, G.; Francois, J. Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol. 2006, 61, 1147–1166. [Google Scholar] [CrossRef] [PubMed]
- Dean, N. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 1995, 92, 1287–1291. [Google Scholar] [CrossRef] [PubMed]
- Ram, A.F.; Klis, F.M. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc. 2006, 1, 2253–2256. [Google Scholar] [CrossRef] [PubMed]
- Roncero, C.; Duran, A. Effect of Calcofluor white and Congo red on fungal cell wall morphogenesis: In vivo activation of chitin polymerization. J. Bacteriol. 1985, 163, 1180–1185. [Google Scholar] [PubMed]
- Girrbach, V.; Zeller, T.; Priesmeier, M.; Strahl-Bolsinger, S. Structure-function analysis of the dolichyl phosphate-mannose: Protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 2000, 275, 19288–19296. [Google Scholar] [CrossRef] [PubMed]
- Zatorska, E.; Strahl, S.; Centre for Organismal Studies, Heidelberg University, Heidelberg, Germany. Immunoblot of HA-tagged Pmt2 probed with polyclonal αPmt2 serum [91]. 2016. [Google Scholar]
- The UniProt Consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. [Google Scholar]
- Bano-Polo, M.; Baldin, F.; Tamborero, S.; Marti-Renom, M.A.; Mingarro, I. N-glycosylation efficiency is determined by the distance to the C-terminus and the amino acid preceding an Asn-Ser-Thr sequon. Protein Sci. 2011, 20, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Zatorska, E.; Strahl, S.; Centre for Organismal Studies, Heidelberg University, Heidelberg, Germany. Immunoblot of whole cell extracts of cycloheximide treated cells (strains EZY50 and EZY53) probed with HA-directed antibody (Covance; Dedham, MA, USA). 2016. [Google Scholar]
- Gentzsch, M.; Tanner, W. Protein-O-glycosylation in yeast: Protein-specific mannosyltransferases. Glycobiology 1997, 7, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Russo, P.; Kalkkinen, N.; Sareneva, H.; Paakkola, J.; Makarow, M. A heat shock gene from Saccharomyces cerevisiae encoding a secretory glycoprotein. Proc. Natl. Acad. Sci. USA 1992, 89, 8857. [Google Scholar] [CrossRef] [PubMed]
- Kuranda, M.J.; Robbins, P.W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 19758–19767. [Google Scholar] [PubMed]
- Manya, H.; Akasaka-Manya, K.; Nakajima, A.; Kawakita, M.; Endo, T. Role of N-glycans in maintaining the activity of protein O-mannosyltransferases POMT1 and POMT2. J. Biochem. 2010, 147, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P. Architecture and biosynthesis of the Saccharomyces cerevisiae cell wall. Genetics 2012, 192, 775–818. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Ng, D.T. Glycosylation-directed quality control of protein folding. Nat. Rev. Mol. Cell Biol. 2015, 16, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Loibl, M.; Strahl, S. Protein O-mannosylation: What we have learned from baker’s yeast. Biochim. Biophys. Acta 2013, 1833, 2438–2446. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, F.; Aebi, M. Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 2011, 21, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Ballou, L.; Hitzeman, R.A.; Lewis, M.S.; Ballou, C.E. Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl. Acad. Sci. USA 1991, 88, 3209–3212. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Nagasu, T.; Shimma, Y.; Kuromitsu, J.; Jigami, Y. OCH1 encodes a novel membrane bound mannosyltransferase: Outer chain elongation of asparagine-linked oligosaccharides. EMBO J. 1992, 11, 2511–2519. [Google Scholar] [PubMed]
- Vossen, J.H.; Muller, W.H.; Lipke, P.N.; Klis, F.M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 1997, 179, 2202–2209. [Google Scholar] [CrossRef] [PubMed]
- Orlean, P.; Menon, A.K. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J. Lipid Res. 2007, 48, 993–1011. [Google Scholar] [CrossRef] [PubMed]
- Castillon, G.A.; Aguilera-Romero, A.; Manzano-Lopez, J.; Epstein, S.; Kajiwara, K.; Funato, K.; Watanabe, R.; Riezman, H.; Muniz, M. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell 2011, 22, 2924–2936. [Google Scholar] [CrossRef] [PubMed]
- Lommel, M.; Bagnat, M.; Strahl, S. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 2004, 24, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Gentzsch, M.; Tanner, W.; Herskowitz, I. O-Glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization, and function in daughter cells. J. Cell Biol. 1999, 145, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Fuller, R.S.; Brake, A.; Thorner, J. Yeast prohormone processing enzyme (KEX2 gene product) is a Ca2+-dependent serine protease. Proc. Natl. Acad. Sci. USA 1989, 86, 1434–1438. [Google Scholar] [CrossRef] [PubMed]
- Proszynski, T.J.; Simons, K.; Bagnat, M. O-glycosylation as a sorting determinant for cell surface delivery in yeast. Mol. Biol. Cell 2004, 15, 1533–1543. [Google Scholar] [CrossRef] [PubMed]
- Lehle, L.; Strahl, S.; Tanner, W. Protein glycosylation, conserved from yeast to man: A model organism helps elucidate congenital human diseases. Angew. Chem. 2006, 45, 6802–6818. [Google Scholar] [CrossRef] [PubMed]
- Lennarz, W.J. Studies on oligosaccharyl transferase in yeast. Acta Biochim. Pol. 2007, 54, 673–677. [Google Scholar] [PubMed]
- Chavan, M.; Yan, A.; Lennarz, W.J. Subunits of the translocon interact with components of the oligosaccharyl transferase complex. J. Biol. Chem. 2005, 280, 22917–22924. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.A.; Lussier, M.; Veronneau, S.; Sdicu, A.M.; Herscovics, A.; Bussey, H. Mnt2p and Mnt3p of Saccharomyces cerevisiae are members of the Mnn1p family of α-1,3-mannosyltransferases responsible for adding the terminal mannose residues of O-linked oligosaccharides. Glycobiology 1999, 9, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; Sdicu, A.M.; Bussey, H. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1999, 1426, 323–334. [Google Scholar] [CrossRef]
- Schulz, B.L.; Stirnimann, C.U.; Grimshaw, J.P.; Brozzo, M.S.; Fritsch, F.; Mohorko, E.; Capitani, G.; Glockshuber, R.; Grutter, M.G.; Aebi, M. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. USA 2009, 106, 11061–11066. [Google Scholar] [CrossRef] [PubMed]
- Knauer, R.; Lehle, L. The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J. Biol. Chem. 1999, 274, 17249–17256. [Google Scholar] [CrossRef] [PubMed]
- Zacchi, L.F.; Schulz, B.L. SWATH-MS Glycoproteomics Reveals Consequences of Defects in the Glycosylation Machinery. Mol. Cell. Proteom. 2016, 15, 2435–2447. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, M.F.; Bailey, U.M.; Tan, N.Y.; Stark, A.P.; Schulz, B.L. Polypeptide binding specificities of Saccharomyces cerevisiae oligosaccharyltransferase accessory proteins Ost3p and Ost6p. Protein Sci. 2011, 20, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Jamaluddin, M.F.; Bailey, U.M.; Schulz, B.L. Oligosaccharyltransferase subunits bind polypeptide substrate to locally enhance N-glycosylation. Mol. Cell. Proteom. 2014, 13, 3286–3293. [Google Scholar] [CrossRef] [PubMed]
- Jung, U.S.; Levin, D.E. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 1999, 34, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Iyer, V.R.; Horak, C.E.; Scafe, C.S.; Botstein, D.; Snyder, M.; Brown, P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 2001, 409, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, H.; Saltsman, K.; Gasch, A.P.; Li, H.X.; Ogawa, N.; Botstein, D.; Brown, P.O.; Cyert, M.S. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002, 277, 31079–31088. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ng, B.S.; Thibault, G. Endoplasmic reticulum stress response in yeast and humans. Biosci. Rep. 2014, 34, e00118. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feldman, D.E.; Deng, C.; Brown, J.A.; De Giacomo, A.F.; Gaw, A.F.; Shi, G.; Le, Q.T.; Brown, J.M.; Koong, A.C. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 2005, 3, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, M.; Cunningham, K.W. Mitogen-activated protein kinase stimulation of Ca2+ signaling is required for survival of endoplasmic reticulum stress in yeast. Mol. Biol. Cell 2003, 14, 4296–4305. [Google Scholar] [CrossRef] [PubMed]
- Gietz, R.D.; Schiestl, R.H.; Willems, A.R.; Woods, R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 1995, 11, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.; Donald, K.A.; Griffiths, D.E. DMSO-enhanced whole cell yeast transformation. Nucleic Acids Res. 1991, 19, 5791. [Google Scholar] [CrossRef] [PubMed]
- Looke, M.; Kristjuhan, K.; Kristjuhan, A. Extraction of genomic DNA from yeasts for PCR-based applications. BioTechniques 2011, 50, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Brachmann, C.B.; Davies, A.; Cost, G.J.; Caputo, E.; Li, J.; Hieter, P.; Boeke, J.D. Designer deletion strains derived from Saccharomyces cerevisiae S288C: Auseful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 1998, 14, 115–132. [Google Scholar] [CrossRef]
- Robinson, J.S.; Klionsky, D.J.; Banta, L.M.; Emr, S.D. Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988, 8, 4936–4948. [Google Scholar] [CrossRef] [PubMed]
- Lussier, M.; Gentzsch, M.; Sdicu, A.M.; Bussey, H.; Tanner, W. Protein O-glycosylation in yeast. The PMT2 gene specifies a second protein O-mannosyltransferase that functions in addition to the PMT1-encoded activity. J. Biol. Chem. 1995, 270, 2770–2775. [Google Scholar] [CrossRef] [PubMed]
- Guldener, U.; Heck, S.; Fielder, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R. PCR Protocols. A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press Inc.: San Diego, CA, USA, 1990; pp. 177–183. [Google Scholar]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Young, B.P.; Loewen, C.J. Balony: A software package for analysis of data generated by synthetic genetic array experiments. BMC Bioinform. 2013, 14, 354. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae. Purification and characterization of the dolichyl-phosphate-D-mannose-protein O-D-mannosyltransferase. Eur. J. Biochem. 1991, 196, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Gentzsch, M.; Immervoll, T.; Tanner, W. Protein O-glycosylation in Saccharomyces cerevisiae: The protein O-mannosyltransferases Pmt1p and Pmt2p function as heterodimer. FEBS Lett. 1995, 377, 128–130. [Google Scholar] [CrossRef]
- Immervoll, T.; Gentzsch, M.; Tanner, W. PMT3 and PMT4, two new members of the protein-O-mannosyltransferase gene family of Saccharomyces cerevisiae. Yeast 1995, 11, 1345–1351. [Google Scholar] [CrossRef] [PubMed]



| GO Term Biological Process | Gene Names |
|---|---|
| GPI anchor biosynthetic process | BST1, CWH43, ERI1, GPI11, GUP1, GWT1, LAS21, MCD4, TED1 |
| Response to osmotic stress | MID2, NBP2, NHA1, RGD1, RVS161, SKN7, STE20, WSC1 |
| ER unfolded protein response | BCK1, HAC1, IRE1, SLT2, VPS74 |
| Fungal-type cell wall organization | ACK1, CHS1, CNB1, CWH43, IRE1, KRE9, MID2, PMT1, PMT2, PMT6, PMT7, SIM1, WSC1 |
| Protein O-linked glycosylation | PGI1, PMT1, PMT2, PMT6, PMT7, VRG4 |
| Protein N-linked glycosylation | ALG6, ALG7, CWH41, OST3, PGI1, ROT2, VRG4, WBP1 |
| Establishment of cell polarity | BCK1, BEM1, BEM2, RGD1, SAC7, SPA2, WSC1 |
| ER-associated misfolded protein catabolic process | PMT1, PMT2, PMT6, PMT7 |
| Cellular cell wall organization | CHS1, CHS3, CHS7, KRE9, MCD4, SIM1, SKT5, TRS65, WSC1 |
| Cell wall integrity pathway | ACK1, BCK1, BEM2, MID2, MKK1, SAC7, SLT2, SPA2, WSC1 |
| Calcineurin signaling pathway | CCH1, CMP2, CNB1 |
| Strain | [3H]Man Incorporation (×103 dpm) ± SD | % ± SD |
|---|---|---|
| Pmt2-HA | 3.909 ± 1.348 | 100 ± 35 |
| pmt2Δ | 0.165 ± 0.05 | 4 ± 1 |
| Pmt2*-HA | 2.328 ± 0.625 | 60 ± 16 |
| Strain | Genotype | Reference/Source |
|---|---|---|
| BY4741 (wild-type) | MATa met15-Δ0 his3-Δ1 leu2-Δ0 ura3-Δ0 | [83] |
| ost3Δ | BY4741 except ost3Δ::kanMX4 | Euroscarf |
| ost6Δ | BY4741 except ost6Δ::kanMX4 | Euroscarf |
| pmt2Δ | BY4741 except pmt2Δ | This study |
| EZY48 | pmt2Δ with pRS415 | This study |
| EZY50 | pmt2Δ with pEZ43 | This study |
| EZY51 | pmt2Δ with pEZ56 | This study |
| EZY52 | pmt2Δ with pEZ57 | This study |
| EZY53 | pmt2Δ with pEZ58 | This study |
| EZY54 | EZY48 with pWX206 | This study |
| EZY55 | EZY50 with pWX206 | This study |
| EZY58 | EZY53 with pWX206 | This study |
| SEY6210 | MATα lys2-801 his3-Δ200 leu2-3,112 trp1-Δ901 ura3-52 suc2-Δ9 | [84] |
| pmt1Δ | SEY6210 except pmt1::HIS3 | [85] |
| EZY66 | SEY6210 except pmt2::LEU2 pmt1-N390A/N513A/N743A-3xHA::kanMX6 and with pRS416 | This study |
| EZY67 | SEY6210 except pmt2::LEU2 pmt1-N390A/N513A/N743A-3xHA::kanMX6 and with pEZ79 | This study |
| EZY68 | SEY6210 except pmt2::LEU2 pmt1-N390A/N513A/N743A-3xHA::kanMX6 and with pEZ78 | This study |
| EZY88 | ost3Δ with pEZ82 | This study |
| JHY1 | SEY6210 except PMT1-3xHA::kanMX6 | [5] |
| MLY67 | SEY6210 except pmt1-N390A/N513A/N743A-3xHA::kanMX6 | [5] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zatorska, E.; Gal, L.; Schmitt, J.; Bausewein, D.; Schuldiner, M.; Strahl, S. Cellular Consequences of Diminished Protein O-Mannosyltransferase Activity in Baker’s Yeast. Int. J. Mol. Sci. 2017, 18, 1226. https://doi.org/10.3390/ijms18061226
Zatorska E, Gal L, Schmitt J, Bausewein D, Schuldiner M, Strahl S. Cellular Consequences of Diminished Protein O-Mannosyltransferase Activity in Baker’s Yeast. International Journal of Molecular Sciences. 2017; 18(6):1226. https://doi.org/10.3390/ijms18061226
Chicago/Turabian StyleZatorska, Ewa, Lihi Gal, Jaro Schmitt, Daniela Bausewein, Maya Schuldiner, and Sabine Strahl. 2017. "Cellular Consequences of Diminished Protein O-Mannosyltransferase Activity in Baker’s Yeast" International Journal of Molecular Sciences 18, no. 6: 1226. https://doi.org/10.3390/ijms18061226