Utilizing Zebrafish Visual Behaviors in Drug Screening for Retinal Degeneration
Abstract
:1. Retinal Degeneration Overview
2. Current and Experimental RD Treatments
3. Zebrafish as a Model System for the Eye
4. Zebrafish for Large-Scale In Vivo Drug Screening
5. Zebrafish Visual Behaviors
5.1. Optokinetic Response (OKR) and the Optomotor Response (OMR)
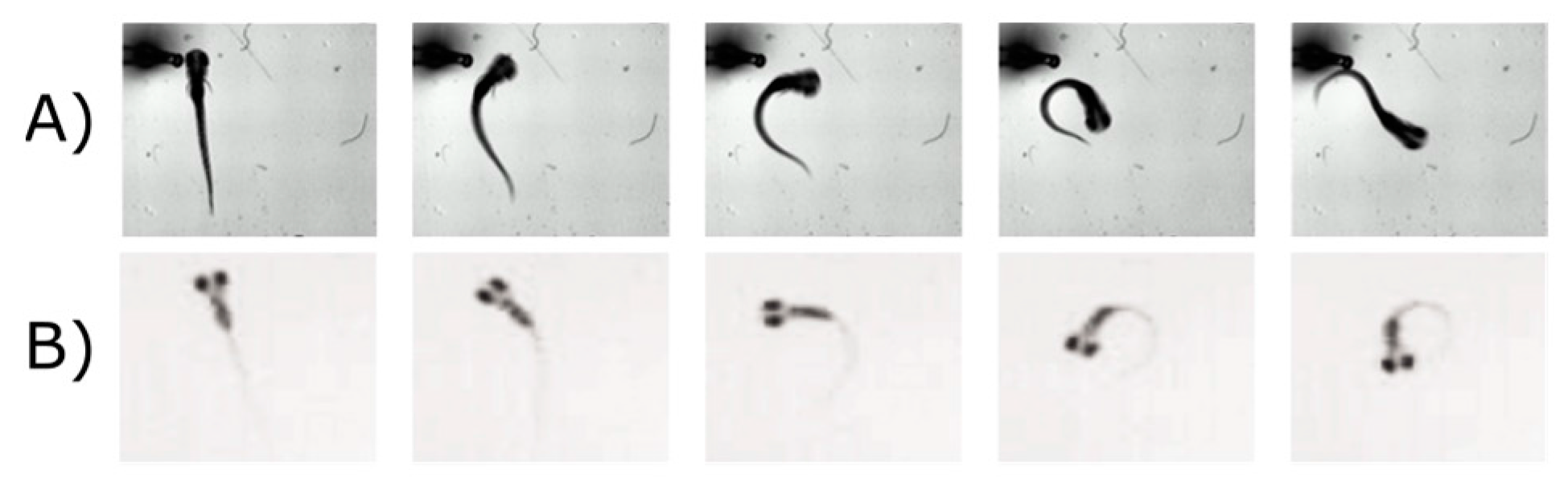
5.2. Visual Motor Response (VMR)
6. Current Issues and Developments of VMR for Drug Discovery
6.1. Extraocular PRs and Locomotor Response
6.2. Neural Basis of VMR
6.3. Rod and Cone Responses in VMR
6.4. VMR Assay Optimization
6.5. VMR of Adult Zebrafish
6.6. Data Analysis of VMR
6.7. Determining Sample Size for VMR Analysis
7. Logistical Considerations in Using Zebrafish for High-Throughput Drug Screening
7.1. Traditional Methods for High-Throughput Drug Screening
7.2. Drawbacks of Using Zebrafish Behavior for Drug Screening
7.3. Auxiliary Technologies to Facilitate High-Throughput Drug Screening in Zebrafish
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Berger, W.; Kloeckener-Gruissem, B.; Neidhardt, J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010, 29, 335–375. [Google Scholar] [CrossRef] [PubMed]
- Hamel, C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006, 1, 40. [Google Scholar] [CrossRef] [PubMed]
- Fadool, J.; Dowling, J. Zebrafish: A model system for the study of eye genetics. Prog. Retin. Eye Res. 2008, 27, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Chung, D.C.; Traboulsi, E.I. Leber congenital amaurosis: Clinical correlations with genotypes, gene therapy trials update, and future directions. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2009, 13, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Weleber, R.G.; Francis, P.J.; Trzupek, K.M.; Beattie, C. Leber Congenital Amaurosis. Available online: http://www.ncbi.nlm.nih.gov/pubmed/20301475 (accessed on 26 January 2017).
- Dryja, T.P.; McGee, T.L.; Reichel, E.; Hahn, L.B.; Cowley, G.S.; Yandell, D.W.; Sandberg, M.A.; Berson, E.L. A point mutation of the rhodopsin gene in one form of retinitis pigmentosa. Nature 1990, 343, 364–366. [Google Scholar] [CrossRef] [PubMed]
- Dryja, T.P.; Hahn, L.B.; Cowley, G.S.; McGee, T.L.; Berson, E.L. Mutation spectrum of the rhodopsin gene among patients with autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1991, 88, 9370–9374. [Google Scholar] [CrossRef] [PubMed]
- Dryja, T.P.; Hahn, L.B.; Reboul, T.; Arnaud, B. Missense mutation in the gene encoding the α subunit of rod transducin in the Nougaret form of congenital stationary night blindness. Nat. Genet. 1996, 13, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Thiadens, A.A.H.J.; den Hollander, A.I.; Roosing, S.; Nabuurs, S.B.; Zekveld-Vroon, R.C.; Collin, R.W.J.; de Baere, E.; Koenekoop, R.K.; van Schooneveld, M.J.; Strom, T.M.; et al. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am. J. Hum. Genet. 2009, 85, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Grau, T.; Dangel, S.; Hurd, R.; Jurklies, B.; Sener, E.C.; Andreasson, S.; Dollfus, H.; Baumann, B.; Bolz, S.; et al. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc. Natl. Acad. Sci. USA 2009, 106, 19581–19586. [Google Scholar] [CrossRef] [PubMed]
- Winick, J.D.; Blundell, M.L.; Galke, B.L.; Salam, A.A.; Leal, S.M.; Karayiorgou, M. Homozygosity mapping of the Achromatopsia locus in the Pingelapese. Am. J. Hum. Genet. 1999, 64, 1679–1685. [Google Scholar] [CrossRef] [PubMed]
- Michaelides, M.; Hardcastle, A.J.; Hunt, D.M.; Moore, A.T. Progressive Cone and Cone-Rod Dystrophies: Phenotypes and Underlying Molecular Genetic Basis. Surv. Ophthalmol. 2006, 51, 232–258. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, A.; Kuppermann, B. Wet age-related macular degeneration. Adv. Drug Deliv. Rev. 2005, 57, 1994–2009. [Google Scholar] [CrossRef] [PubMed]
- Raz-Prag, D.; Zeng, Y.; Sieving, P.A.; Bush, R.A. Photoreceptor protection by adeno-associated virus-mediated LEDGF expression in the RCS rat model of retinal degeneration: Probing the mechanism. Investig. Ophthalmol. Vis. Sci. 2009, 50, 3897–3906. [Google Scholar] [CrossRef] [PubMed]
- Chadderton, N.; Millington-Ward, S.; Palfi, A.; O’Reilly, M.; Tuohy, G.; Humphries, M.M.; Li, T.; Humphries, P.; Kenna, P.F.; Farrar, G.J. Improved Retinal Function in a Mouse Model of Dominant Retinitis Pigmentosa Following AAV-delivered Gene Therapy. Mol. Ther. 2009, 17, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Palfi, A.; Millington-Ward, S.; Chadderton, N.; O’Reilly, M.; Goldmann, T.; Humphries, M.M.; Li, T.; Wolfrum, U.; Humphries, P.; Kenna, P.F.; et al. Adeno-Associated Virus-Mediated Rhodopsin Replacement Provides Therapeutic Benefit in Mice with a Targeted Disruption of the Rhodopsin Gene. Hum. Gene Ther. 2010, 21, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Rowland, T.J.; Buchholz, D.E.; Clegg, D.O. Pluripotent human stem cells for the treatment of retinal disease. J. Cell. Physiol. 2012, 227, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Bermingham-McDonogh, O.; Reh, T.A. Regulated reprogramming in the regeneration of sensory receptor cells. Neuron 2011, 71, 389–405. [Google Scholar] [CrossRef] [PubMed]
- Comyn, O.; Lee, E.; MacLaren, R.E. Induced pluripotent stem cell therapies for retinal disease. Curr. Opin. Neurol. 2010, 23, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.S.; Tezel, T.H.; del Priore, L.V.; Kaplan, H.J. Photoreceptor transplantation in retinitis pigmentosa: Short-term follow-up. Ophthalmology 2003, 110, 383–391. [Google Scholar] [CrossRef]
- Del Bene, F.; Wyart, C. Optogenetics: A new enlightenment age for zebrafish neurobiology. Dev. Neurobiol. 2012, 72, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, B.S.; Marc, R.E.; Bernstein, P.S. Optogenetics for retinal disorders. J. Ophthalmic Vis. Res. 2014, 9, 374–382. [Google Scholar] [PubMed]
- Duncan, J.L.; Richards, T.P.; Arditi, A.; da Cruz, L.; Dagnelie, G.; Dorn, J.D.; Ho, A.C.; Olmos de Koo, L.C.; Barale, P.-O.; Stanga, P.E.; et al. Improvements in vision-related quality of life in blind patients implanted with the Argus II Epiretinal Prosthesis. Clin. Exp. Optom. 2017, 100, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.C.; Humayun, M.S.; Dorn, J.D.; da Cruz, L.; Dagnelie, G.; Handa, J.; Barale, P.-O.; Sahel, J.-A.; Stanga, P.E.; Hafezi, F.; et al. Long-Term Results from an Epiretinal Prosthesis to Restore Sight to the Blind. Ophthalmology 2015, 122, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Oveson, B.C.; Jo, Y.; Lauer, T.W.; Usui, S.; Komeima, K.; Xie, B.; Campochiaro, P.A. Increased expression of glutathione peroxidase 4 strongly protects retina from oxidative damage. Antioxid. Redox Signal. 2009, 11, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Usui, S.; Komeima, K.; Lee, S.Y.; Jo, Y.-J.; Ueno, S.; Rogers, B.S.; Wu, Z.; Shen, J.; Lu, L.; Oveson, B.C.; et al. Increased expression of catalase and superoxide dismutase 2 reduces cone cell death in retinitis pigmentosa. Mol. Ther. 2009, 17, 778–786. [Google Scholar] [CrossRef] [PubMed]
- MacLaren, R.E.; Pearson, R.A.; MacNeil, A.; Douglas, R.H.; Salt, T.E.; Akimoto, M.; Swaroop, A.; Sowden, J.C.; Ali, R.R. Retinal repair by transplantation of photoreceptor precursors. Nature 2006, 444, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.; de Luca, M.; Arsenijevic, Y. Towards therapeutic application of ocular stem cells. Semin. Cell Dev. Biol. 2007, 18, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Enzmann, V.; Ildstad, S.T. Stem Cell-Based Therapeutic Applications in Retinal Degenerative Diseases. Stem Cell Rev. Rep. 2011, 7, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Reardon, S.; Cyranoski, D. Japan stem-cell trial stirs envy. Nature 2014, 513, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.D.; Regillo, C.D.; Lam, B.L.; Eliott, D.; Rosenfeld, P.J.; Gregori, N.Z.; Hubschman, J.-P.; Davis, J.L.; Heilwell, G.; Spirn, M.; et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt’s macular dystrophy: Follow-up of two open-label phase 1/2 studies. Lancet 2015, 385, 509–516. [Google Scholar] [CrossRef]
- Takahashi, M.; Kurimoto, Y. Pilot Safety Study of iPSC-Based Intervention for Wet-Type AMD. Available online: http://www.riken-ibri.jp/AMD/english/summary.html (accessed on 15 Febrary 2017).
- Lin, B.; Koizumi, A.; Tanaka, N.; Panda, S.; Masland, R.H. Restoration of visual function in retinal degeneration mice by ectopic expression of melanopsin. Proc. Natl. Acad. Sci. USA 2008, 105, 16009–16014. [Google Scholar] [CrossRef] [PubMed]
- Lagali, P.S.; Balya, D.; Awatramani, G.B.; Münch, T.A.; Kim, D.S.; Busskamp, V.; Cepko, C.L.; Roska, B. Light-activated channels targeted to ON bipolar cells restore visual function in retinal degeneration. Nat. Neurosci. 2008, 11, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Doroudchi, M.M.; Greenberg, K.P.; Liu, J.; Silka, K.A.; Boyden, E.S.; Lockridge, J.A.; Arman, A.C.; Janani, R.; Boye, S.E.; Boye, S.L.; et al. Virally delivered channelrhodopsin-2 safely and effectively restores visual function in multiple mouse models of blindness. Mol. Ther. 2011, 19, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, A.; Borgonovi, E.; Taylor, R.S.; Sahel, J.-A.; Rizzo, S.; Stanga, P.E.; Kukreja, A.; Walter, P. The cost-effectiveness of the Argus II retinal prosthesis in retinitis pigmentosa patients. BMC Ophthalmol. 2014, 14, 49. [Google Scholar] [CrossRef] [PubMed]
- Berson, E.L.; Rosner, B.; Sandberg, M.A.; Weigel-DiFranco, C.; Willett, W.C. ω-3 Intake and visual acuity in patients with retinitis pigmentosa receiving vitamin A. Arch. Ophthalmol. 2012, 130, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Berson, E.L.; Rosner, B.; Sandberg, M.A.; Hayes, K.C.; Nicholson, B.W.; Weigel-DiFranco, C.; Willett, W. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch. Ophthalmol. 1993, 111, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, D.R.; Locke, K.G.; Wheaton, D.H.; Fish, G.E.; Spencer, R.; Birch, D.G. A randomized, placebo-controlled clinical trial of docosahexaenoic acid supplementation for X-linked retinitis pigmentosa. Am. J. Ophthalmol. 2004, 137, 704–718. [Google Scholar] [PubMed]
- Schaefer, E.J.; Robins, S.J.; Patton, G.M.; Sandberg, M.A.; Weigel-DiFranco, C.A.; Rosner, B.; Berson, E.L. Red blood cell membrane phosphatidylethanolamine fatty acid content in various forms of retinitis pigmentosa. J. Lipid Res. 1995, 36, 1427–1433. [Google Scholar] [PubMed]
- Komeima, K.; Rogers, B.S.; Lu, L.; Campochiaro, P.A. Antioxidants reduce cone cell death in a model of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2006, 103, 11300–11305. [Google Scholar] [CrossRef] [PubMed]
- Komeima, K.; Rogers, B.S.; Campochiaro, P.A. Antioxidants slow photoreceptor cell death in mouse models of retinitis pigmentosa. J. Cell. Physiol. 2007, 213, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Slijkerman, R.W.N.; Song, F.; Astuti, G.D.N.; Huynen, M.A.; van Wijk, E.; Stieger, K.; Collin, R.W.J. The pros and cons of vertebrate animal models for functional and therapeutic research on inherited retinal dystrophies. Prog. Retin. Eye Res. 2015, 48, 137–159. [Google Scholar] [CrossRef] [PubMed]
- Patton, E.E.; Zon, L.I. The art and design of genetic screens: Zebrafish. Nat. Rev. Genet. 2001, 2, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Bilotta, J.; Saszik, S.; Sutherland, S.E. Rod contributions to the electroretinogram of the dark-adapted developing zebrafish. Dev. Dyn. 2001, 222, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.; Schmitt, E.A.; Hárosi, F.I.; Reece, R.J.; Dowling, J.E. Zebrafish ultraviolet visual pigment: Absorption spectrum, sequence, and localization. Proc. Natl. Acad. Sci. USA 1993, 90, 6009–6012. [Google Scholar] [CrossRef] [PubMed]
- Fadool, J.M. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev. Biol. 2003, 258, 277–290. [Google Scholar] [CrossRef]
- Salbreux, G.; Barthel, L.K.; Raymond, P.A.; Lubensky, D.K. Coupling Mechanical Deformations and Planar Cell Polarity to Create Regular Patterns in the Zebrafish Retina. PLoS Comput. Biol. 2012, 8, e1002618. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.A.; Carney, L.H. Cell mosaic patterns in the native and regenerated inner retina of zebrafish: Implications for retinal assembly. J. Comp. Neurol. 2000, 416, 356–367. [Google Scholar] [CrossRef]
- Purves, D.; Augustine, G.; Fitzpatrick, D. Anatomical Distribution of Rods and Cones. Available online: https://www.ncbi.nlm.nih.gov/books/NBK10848/ (accessed on 17 February 2017).
- Jonas, J.B.; Schneider, U.; Naumann, G.O. Count and density of human retinal photoreceptors. Graefes Arch. Clin. Exp. Ophthalmol. 1992, 230, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bibliowicz, J.; Tittle, R.K.; Gross, J.M. Toward a better understanding of human eye disease insights from the zebrafish, Danio rerio. Prog. Mol. Biol. Transl. Sci. 2011, 100, 287–330. [Google Scholar] [PubMed]
- Link, B.A.; Collery, R.F. Zebrafish Models of Retinal Disease. Annu. Rev. Vis. Sci. 2015, 1, 125–153. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.C. The genetics of ocular disorders: Insights from the zebrafish. Birth Defect. Res. Part C Embryo Today Rev. 2011, 93, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.M.; Perkins, B.D. Zebrafish mutants as models for congenital ocular disorders in humans. Mol. Reprod. Dev. 2008, 75, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Brockerhoff, S.E.; Fadool, J.M. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell. Mol. Life Sci. 2011, 68, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, M.; Malicki, J. Genetics of photoreceptor development and function in zebrafish. Int. J. Dev. Biol. 2004, 48, 925–934. [Google Scholar] [CrossRef] [PubMed]
- Nakao, T.; Tsujikawa, M.; Notomi, S.; Ikeda, Y.; Nishida, K. The Role of Mislocalized Phototransduction in Photoreceptor Cell Death of Retinitis Pigmentosa. PLoS ONE 2012, 7, e32472. [Google Scholar] [CrossRef] [PubMed]
- Stearns, G.; Evangelista, M.; Fadool, J.M.; Brockerhoff, S.E. A Mutation in the Cone-Specific pde6 Gene Causes Rapid Cone Photoreceptor Degeneration in Zebrafish. J. Neurosci. 2007, 27, 13866–13874. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.; Williams, P.; Lawrence, O.; Wong, R.O.L.; Brockerhoff, S.E. Wild-Type Cone Photoreceptors Persist Despite Neighboring Mutant Cone Degeneration. J. Neurosci. 2010, 30, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Hawes, N.L.; Hurd, R.E.; Davisson, M.T.; Nusinowitz, S.; Heckenlively, J.R. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef]
- Owens, K.N.; Santos, F.; Roberts, B.; Linbo, T.; Coffin, A.B.; Knisely, A.J.; Simon, J.A.; Rubel, E.W.; Raible, D.W. Identification of genetic and chemical modulators of zebrafish mechanosensory hair cell death. PLoS Genet. 2008, 4, e1000020. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.T.; Link, B.A.; Dowling, J.E.; Schreiber, S.L. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc. Natl. Acad. Sci. USA 2000, 97, 12965–12969. [Google Scholar] [CrossRef] [PubMed]
- Peterson, R.T.; Shaw, S.Y.; Peterson, T.A.; Milan, D.J.; Zhong, T.P.; Schreiber, S.L.; MacRae, C.A.; Fishman, M.C. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat. Biotechnol. 2004, 22, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Novodvorsky, P.; Da Costa, M.M.J.; Chico, T.J.A. Zebrafish-based small molecule screens for novel cardiovascular drugs. Drug Discov. Today Technol. 2013, 10, e109–e114. [Google Scholar] [CrossRef] [PubMed]
- Rihel, J.; Prober, D.A.; Arvanites, A.; Lam, K.; Zimmerman, S.; Jang, S.; Haggarty, S.J.; Kokel, D.; Rubin, L.L.; Peterson, R.T.; et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science 2010, 327, 348–351. [Google Scholar] [CrossRef] [PubMed]
- Kokel, D.; Bryan, J.; Laggner, C.; White, R.; Cheung, C.Y.J.; Mateus, R.; Healey, D.; Kim, S.; Werdich, A.A.; Haggarty, S.J.; et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat. Chem. Biol. 2010, 6, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Bruni, G.; Rennekamp, A.J.; Velenich, A.; McCarroll, M.; Gendelev, L.; Fertsch, E.; Taylor, J.; Lakhani, P.; Lensen, D.; Evron, T.; et al. Zebrafish behavioral profiling identifies multitarget antipsychotic-like compounds. Nat. Chem. Biol. 2016, 12, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Dinday, M.T.; Baraban, S.C. Large-Scale Phenotype-Based Antiepileptic Drug Screening in a Zebrafish Model of Dravet Syndrome. eNeuro 2015, 2. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, V.E.; Varshney, G.K.; Lee, M.; Bupp, S.; Xu, L.; Shinn, P.; Crawford, N.P.; Inglese, J.; Burgess, S.M. Phenotype-driven chemical screening in zebrafish for compounds that inhibit collective cell migration identifies multiple pathways potentially involved in metastatic invasion. Dis. Model. Mech. 2015, 8, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rhee, D.K.; Malhotra, R.; Mayeur, C.; Hurst, L.A.; Ager, E.; Shelton, G.; Kramer, Y.; McCulloh, D.; Keefe, D.; et al. Progesterone receptor membrane component-1 regulates hepcidin biosynthesis. J. Clin. Investig. 2015, 126, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.K.; Ryu, J.H.; Jin, Y.N.; Roberts, L.D.; Dejam, A.; Gerszten, R.E.; Peterson, R.T. PTPMT1 inhibition lowers glucose through succinate dehydrogenase phosphorylation. Cell Rep. 2016, 10, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Asnani, A.; Zou, L.; Bentley, V.L.; Yu, M.; Wang, Y.; Dellaire, G.; Sarkar, K.S.; Dai, M.; Chen, H.H.; et al. Visnagin protects against doxorubicin-induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase. Sci. Transl. Med. 2014, 6, 266ra170. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Sarkar, K.S.; Jin, Y.N.; Liu, Y.; Kokel, D.; Van Ham, T.J.; Roberts, L.D.; Gerszten, R.E.; MacRae, C.A.; Peterson, R.T. An in vivo zebrafish screen identifies organophosphate antidotes with diverse mechanisms of action. J. Biomol. Screen. 2013, 18, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Kokel, D.; Cheung, C.Y.J.; Mills, R.; Coutinho-Budd, J.; Huang, L.; Setola, V.; Sprague, J.; Jin, S.; Jin, Y.N.; Huang, X.-P.; et al. Photochemical activation of TRPA1 channels in neurons and animals. Nat. Chem. Biol. 2013, 9, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.K.; Roberts, L.D.; Liu, Y.; Mahon, S.B.; Kim, S.; Ryu, J.H.; Werdich, A.; Januzzi, J.L.; Boss, G.R.; Rockwood, G.A.; et al. Chemical and metabolomic screens identify novel biomarkers and antidotes for cyanide exposure. FASEB J. 2013, 27, 1928–1938. [Google Scholar] [CrossRef] [PubMed]
- Baxendale, S.; Holdsworth, C.J.; Meza Santoscoy, P.L.; Harrison, M.R.M.; Fox, J.; Parkin, C.A.; Ingham, P.W.; Cunliffe, V.T. Identification of compounds with anti-convulsant properties in a zebrafish model of epileptic seizures. Dis. Model. Mech. 2012, 5, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Emran, F.; Rihel, J.; Adolph, A.R.; Wong, K.Y.; Kraves, S.; Dowling, J.E. OFF ganglion cells cannot drive the optokinetic reflex in zebrafish. Proc. Natl. Acad. Sci. USA 2007, 104, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Emran, F.; Rihel, J.; Dowling, J.E. A behavioral assay to measure responsiveness of zebrafish to changes in light intensities. J. Vis. Exp. 2008, e923. [Google Scholar] [CrossRef] [PubMed]
- Neuhauss, S.C.; Biehlmaier, O.; Seeliger, M.W.; Das, T.; Kohler, K.; Harris, W.A.; Baier, H. Genetic disorders of vision revealed by a behavioral screen of 400 essential loci in zebrafish. J. Neurosci. 1999, 19, 8603–8615. [Google Scholar] [PubMed]
- Muto, A.; Orger, M.B.; Wehman, A.M.; Smear, M.C.; Kay, J.N.; Page-McCaw, P.S.; Gahtan, E.; Xiao, T.; Nevin, L.M.; Gosse, N.J.; et al. Forward genetic analysis of visual behavior in zebrafish. PLoS Genet. 2005, 1, e66. [Google Scholar] [CrossRef] [PubMed]
- Bilotta, J. Effects of abnormal lighting on the development of zebrafish visual behavior. Behav. Brain Res. 2000, 116, 81–87. [Google Scholar] [CrossRef]
- Brockerhoff, S.E.; Hurley, J.B.; Janssen-Bienhold, U.; Neuhauss, S.C.; Driever, W.; Dowling, J.E. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc. Natl. Acad. Sci. USA 1995, 92, 10545–10549. [Google Scholar] [CrossRef] [PubMed]
- Chhetri, J.; Jacobson, G.; Gueven, N. Zebrafish—on the move towards ophthalmological research. Eye 2014, 28, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Rihel, J.; Schier, A.F. Behavioral screening for neuroactive drugs in zebrafish. Dev. Neurobiol. 2012, 72, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Portugues, R.; Engert, F. The neural basis of visual behaviors in the larval zebrafish. Curr. Opin. Neurobiol. 2009, 19, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Orger, M.B.; Gahtan, E.; Muto, A.; Page-McCaw, P.; Smear, M.C.; Baier, H. Behavioral screening assays in zebrafish. Methods Cell Biol. 2004, 77, 53–68. [Google Scholar] [PubMed]
- Brockerhoff, S.E. Measuring the optokinetic response of zebrafish larvae. Nat. Protoc. 2006, 1, 2448–2451. [Google Scholar] [CrossRef] [PubMed]
- Brockerhoff, S.E.; Hurley, J.B.; Niemi, G.A.; Dowling, J.E. A new form of inherited red-blindness identified in zebrafish. J. Neurosci. 1997, 17, 4236–4242. [Google Scholar] [PubMed]
- Easter, S.S.; Gregory Nicola, J.N. The Development of Eye Movements in the Zebrafish (Danio rerio). Dev. Psychobiol. 1997, 31, 267–276. [Google Scholar] [CrossRef]
- Baier, H.; Orger, M.B.; Smear, M.C.; Anstis, S.M. Perception of Fourier and non-Fourier motion by larval zebrafish. Nat. Neurosci. 2000, 3, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.M.; Perkins, B.D.; Amsterdam, A.; Egaña, A.; Darland, T.; Matsui, J.I.; Sciascia, S.; Hopkins, N.; Dowling, J.E. Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics 2005, 170, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.R.; Hurley, J.B.; van Epps, H.A.; Brockerhoff, S.E. A zebrafish model for pyruvate dehydrogenase deficiency: Rescue of neurological dysfunction and embryonic lethality using a ketogenic diet. Proc. Natl. Acad. Sci. USA 2004, 101, 4584–4589. [Google Scholar] [CrossRef] [PubMed]
- Roosing, S.; Lamers, I.J.C.; de Vrieze, E.; van den Born, L.I.; Lambertus, S.; Arts, H.H.; POC1B Study Group; Peters, T.A.; Hoyng, C.B.; Kremer, H.; et al. Disruption of the basal body protein POC1B results in autosomal-recessive cone-rod dystrophy. Am. J. Hum. Genet. 2014, 95, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Chen, S.; Liu, S.; Zhang, C.; Peng, G. Effects of lorazepam and WAY-200070 in larval zebrafish light/dark choice test. Neuropharmacology 2015, 95, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Deeti, S.; O’Farrell, S.; Kennedy, B.N. Early safety assessment of human oculotoxic drugs using the zebrafish visualmotor response. J. Pharmacol. Toxicol. Methods 2014, 69, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Richards, F.M.; Alderton, W.K.; Kimber, G.M.; Liu, Z.; Strang, I.; Redfern, W.S.; Valentin, J.-P.; Winter, M.J.; Hutchinson, T.H. Validation of the use of zebrafish larvae in visual safety assessment. J. Pharmacol. Toxicol. Methods 2008, 58, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-Y.; Neuhauss, S.C.F. The optokinetic response in zebrafish and its applications. Front. Biosci. 2008, 13, 1899–1916. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, G.; Jelfs, B.; Carmer, R.; Venkatraman, P.; Ghadami, M.; Brown, S.A.; Pang, C.P.; Leung, Y.F.; Chan, R.H.M.; et al. Computational classification of different wild-type zebrafish strains based on their variation in light-induced locomotor response. Comput. Biol. Med. 2016, 69, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Carmer, R.; Zhang, G.; Venkatraman, P.; Brown, S.A.; Pang, C.-P.; Zhang, M.; Ma, P.; Leung, Y.F. Statistical analysis of zebrafish locomotor response. PLoS ONE 2015, 10, e0139521. [Google Scholar] [CrossRef] [PubMed]
- Viewpoint LifeSciences. Available online: http://csl.mendeley.com/styles/485647231/MDPI-LOGAN-FINAL (accessed on 23 May 2017).
- Noldus. Available online: http://www.noldus.com (accessed on 23 May 2017).
- Zhou, Y.; Cattley, R.T.; Cario, C.L.; Bai, Q.; Burton, E.A. Quantification of larval zebrafish motor function in multiwell plates using open-source MATLAB applications. Nat. Protoc. 2014, 9, 1533–1548. [Google Scholar] [CrossRef] [PubMed]
- Maurer, C.M.; Schonthaler, H.B.; Mueller, K.P.; Neuhauss, S.C.F. Distinct Retinal Deficits in a Zebrafish Pyruvate Dehydrogenase-Deficient Mutant. J. Neurosci. 2010, 30, 11962–11972. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiang, L.; Liu, Y.; Venkatraman, P.; Chong, L.; Cho, J.; Bonilla, S.; Jin, Z.-B.; Pang, C.P.; Ko, K.M.; et al. A naturally-derived compound schisandrin B enhanced light sensation in the pde6c zebrafish model of retinal degeneration. PLoS ONE 2016, 11, e0149663. [Google Scholar] [CrossRef] [PubMed]
- Ingebretson, J.J.; Masino, M.A. Quantification of locomotor activity in larval zebrafish: Considerations for the design of high-throughput behavioral studies. Front. Neural Circuits 2013, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Colwill, R.M.; Creton, R. Locomotor behaviors in zebrafish (Danio rerio) larvae. Behav. Processes 2011, 86, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chan, R.H.M.; Chow, T.W.S.; Zhang, L.; Bonilla, S.; Pang, C.-P.; Zhang, M.; Leung, Y.F. A high-throughput zebrafish screening method for visual mutants by light-induced locomotor response. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2014, 11, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Champagne, D.L.; Richardson, M.K. Behavioral profiling of zebrafish embryos exposed to a panel of 60 water-soluble compounds. Behav. Brain Res. 2012, 228, 272–283. [Google Scholar] [CrossRef] [PubMed]
- De Esch, C.; van der Linde, H.; Slieker, R.; Willemsen, R.; Wolterbeek, A.; Woutersen, R.; et al. Locomotor activity assay in zebrafish larvae: Influence of age, strain and ethanol. Neurotoxicol. Teratol. 2012, 34, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mil, H.G.J.v.; Richardson, M.K. Large-Scale Assessment of the Zebrafish Embryo as a Possible Predictive Model in Toxicity Testing. PLoS ONE 2011, 6, e21076. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.; Neuzeret, F.; Fabreges, B.; Froc, C.; Bedu, S.; Bally-Cuif, L.; Norton, W.H.J. Inter-individual and inter-strain variations in zebrafish locomotor ontogeny. PLoS ONE 2013, 8, e70172. [Google Scholar] [CrossRef] [PubMed]
- Vignet, C.; Bégout, M.-L.; Péan, S.; Lyphout, L.; Leguay, D.; Cousin, X. Systematic Screening of Behavioral Responses in Two Zebrafish Strains. Zebrafish 2013, 10, 365–375. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.; Hunter, D.L.; Padnos, B.; Frady, S.; MacPhail, R.C. Assessing locomotor activity in larval zebrafish: Influence of extrinsic and intrinsic variables. Neurotoxicol. Teratol. 2011, 33, 624–630. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.M.; Fero, K.; Arrenberg, A.B.; Bergeron, S.A.; Driever, W.; Burgess, H.A. Deep Brain Photoreceptors Control Light-Seeking Behavior in Zebrafish Larvae. Curr. Biol. 2012, 22, 2042–2047. [Google Scholar] [CrossRef] [PubMed]
- Beker van Woudenberg, A.; Wolterbeek, A.; te Brake, L.; Snel, C.; Menke, A.; Rubingh, C.; de Groot, D.; Kroese, D. A category approach to predicting the developmental (neuro) toxicity of organotin compounds: The value of the zebrafish (Danio rerio) embryotoxicity test (ZET). Reprod. Toxicol. 2013, 41, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Conklin, E.E.; Lee, K.L.; Schlabach, S.A.; Woods, I.G. VideoHacking: Automated Tracking and Quantification of Locomotor Behavior with Open Source Software and Off-the-Shelf Video Equipment. J. Undergrad. Neurosci. Educ. 2015, 13, A120–A125. [Google Scholar] [PubMed]
- Pittman, J.T.; Ichikawa, K.M. iPhone® applications as versatile video tracking tools to analyze behavior in zebrafish (Danio rerio). Pharmacol. Biochem. Behav. 2013, 106, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.P.; Neuhauss, S.C.F. Automated visual choice discrimination learning in zebrafish (Danio rerio). J. Integr. Neurosci. 2012, 11, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.P.; Neuhauss, S.C.F. Sunscreen for Fish: Co-Option of UV Light Protection for Camouflage. PLoS ONE 2014, 9, e87372. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.A.; Granato, M. Sensorimotor Gating in Larval Zebrafish. J. Neurosci. 2007, 27, 4984–4994. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: http://www.r-project.org (accessed on 15 May 2017).
- Metcalfe, W.K.; Mendelson, B.; Kimmel, C.B. Segmental homologies among reticulospinal neurons in the hindbrain of the zebrafish larva. J. Comp. Neurol. 1986, 251, 147–159. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.A.; Granato, M. Modulation of locomotor activity in larval zebrafish during light adaptation. J. Exp. Biol. 2007, 210, 2526–2539. [Google Scholar] [CrossRef] [PubMed]
- Eaton, R.C.; Bombardieri, R.A.; Meyer, D.L. The Mauthner-initiated startle response in teleost fish. J. Exp. Biol. 1977, 66, 65–81. [Google Scholar] [PubMed]
- Liu, K.S.; Fetcho, J.R. Laser ablations reveal functional relationships of segmental hindbrain neurons in zebrafish. Neuron 1999, 23, 325–335. [Google Scholar] [CrossRef]
- Lorent, K.; Liu, K.S.; Fetcho, J.R.; Granato, M. The zebrafish space cadet gene controls axonal pathfinding of neurons that modulate fast turning movements. Development 2001, 128, 2131–2142. [Google Scholar] [PubMed]
- Moyano, M.; Porteros, Á.; Dowling, J.E. The effects of nicotine on cone and rod b-wave responses in larval zebrafish. Vis. Neurosci. 2013, 30, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, P.; Carmer, R.; Pang, C.-P.; Zhang, M.; Leung, Y.F. Understanding the contribution of photoreceptors to the Visual Motor Response. Investig. Ophthalmol. Vis. Sci. 2015, 56, 998. [Google Scholar]
- Burton, C.E.; Zhou, Y.; Bai, Q.; Burton, E.A. Spectral properties of the zebrafish visual motor response. Neurosci. Lett. 2017, 646, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Mora-Zamorano, F.X.; Klingler, R.; Murphy, C.A.; Basu, N.; Head, J.; Carvan, M.J. Parental Whole Life Cycle Exposure to Dietary Methylmercury in Zebrafish (Danio rerio) Affects the Behavior of Offspring. Environ. Sci. Technol. 2016, 50, 4808–4816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chong, L.; Cho, J.; Liao, P.-C.; Shen, F.; Leung, Y.F. Drug Screening to Treat Early-Onset Eye Diseases. Asia Pac. J. Ophthalmol. 2012, 1, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Escudero, A.; Vicente-Page, J.; Hinz, R.C.; Arganda, S.; de Polavieja, G.G. idTracker: Tracking individuals in a group by automatic identification of unmarked animals. Nat. Methods 2014, 11, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, P.; Cassidy, P.; Carmer, R.; Zhang, G.; Venkatraman, P.; Brown, S.A.; Pang, C.-P.; Zhong, W.; Zhang, M.; et al. Statistical analysis of zebrafish locomotor behaviour by logistic generalized linear mixed models. Sci. Rep. 2017, in press. [Google Scholar] [CrossRef]
- Rihel, J.; Prober, D.A.; Schier, A.F. Monitoring Sleep and Arousal in Zebrafish. Methods Cell Biol. 2010, 100, 281–294. [Google Scholar] [PubMed]
- Scott, C.A.; Marsden, A.N.; Slusarski, D.C. Automated, high-throughput, in vivo analysis of visual function using the zebrafish. Dev. Dyn. 2016, 245, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Michelini, E.; Cevenini, L.; Mezzanotte, L.; Coppa, A.; Roda, A. Cell-based assays: Fuelling drug discovery. Anal. Bioanal. Chem. 2010, 398, 227–238. [Google Scholar] [CrossRef] [PubMed]
- An, W.F.; Tolliday, N. Cell-Based Assays for High-Throughput Screening. Mol. Biotechnol. 2010, 45, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.; Rees, S. Cell-Based Versus Isolated Target Screening: How Lucky Do You Feel? J. Biomol. Screen 2001, 6, 69–74. [Google Scholar] [CrossRef] [PubMed]
- Szymański, P.; Markowicz, M.; Mikiciuk-Olasik, E. Adaptation of High-Throughput Screening in Drug Discovery—Toxicological Screening Tests. Int. J. Mol. Sci. 2011, 13, 427–452. [Google Scholar] [CrossRef] [PubMed]
- Kasibhatla, S.; Gourdeau, H.; Meerovitch, K.; Drewe, J.; Reddy, S.; Qiu, L.; Zhang, H.; Bergeron, F.; Bouffard, D.; Yang, Q.; et al. Discovery and mechanism of action of a novel series of apoptosis inducers with potential vascular targeting activity. Mol. Cancer Ther. 2004, 3, 1365–1374. [Google Scholar] [PubMed]
- Mueller, H.; Kassack, M.U.; Wiese, M. Comparison of the Usefulness of the MTT, ATP, and Calcein Assays to Predict the Potency of Cytotoxic Agents in Various Human Cancer Cell Lines. J. Biomol. Screen 2004, 9, 506–515. [Google Scholar] [CrossRef] [PubMed]
- Drews, J. Drug discovery: A historical perspective. Science 2000, 287, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Samsdodd, F. Target-based drug discovery: Is something wrong? Drug Discov. Today 2005, 10, 139–147. [Google Scholar] [CrossRef]
- Terstappen, G.C.; Schlüpen, C.; Raggiaschi, R.; Gaviraghi, G. Target deconvolution strategies in drug discovery. Nat. Rev. Drug Discov. 2007, 6, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Swinney, D.C. Phenotypic vs. Target-Based Drug Discovery for First-in-Class Medicines. Clin. Pharmacol. Ther. 2013, 93, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Swinney, D.C.; Anthony, J. How were new medicines discovered? Nat. Rev. Drug Discov. 2011, 10, 507–519. [Google Scholar] [CrossRef] [PubMed]
- Gower, A.J.; Noyer, M.; Verloes, R.; Gobert, J.; Wülfert, E. UCB L059, a novel anti-convulsant drug: Pharmacological profile in animals. Eur. J. Pharmacol. 1992, 222, 193–203. [Google Scholar] [CrossRef]
- Emran, F.; Rihel, J.; Adolph, A.R.; Dowling, J.E. Zebrafish larvae lose vision at night. Proc. Natl. Acad. Sci. USA 2010, 107, 6034–6039. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.H.; Dobrinski, K.P.; Lee, A.S.; Gokcumen, O.; Mills, R.E.; Shi, X.; Chong, W.W.S.; Chen, J.Y.H.; Yoo, P.; David, S.; et al. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Natl. Acad. Sci. USA 2012, 109, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Coe, T.S.; Hamilton, P.B.; Griffiths, A.M.; Hodgson, D.J.; Wahab, M.A.; Tyler, C.R. Genetic variation in strains of zebrafish (Danio rerio) and the implications for ecotoxicology studies. Ecotoxicology 2009, 18, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Cross, L.M.; Cook, M.A.; Lin, S.; Chen, J.-N.; Rubinstein, A.L. Rapid Analysis of Angiogenesis Drugs in a Live Fluorescent Zebrafish Assay. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 911–912. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wan, X.; Li, Y.; Zhou, W.; Peng, L. Multiplexed 3D FRET imaging in deep tissue of live embryos. Sci. Rep. 2015, 5, 13991. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Ramel, M.-C.; Kumar, S.; Alexandrov, Y.; Kelly, D.J.; Warren, S.C.; Kerry, L.; Lockwood, N.; Frolov, A.; Frankel, P.; et al. Visualising apoptosis in live zebrafish using fluorescence lifetime imaging with optical projection tomography to map FRET biosensor activity in space and time. J. Biophotonics 2016, 9, 414–424. [Google Scholar] [CrossRef] [PubMed]
- Pentair Aquatic Eco-Systems. Available online: http://pentairaes.com (accessed on 23 May 2017).
- Techniplast. Available online: www.tecniplast.it (accessed on 23 May 2017).
- Aquaneering Incorporated. Available online: http://www.aquaneering.com (accessed on 23 May 2017).
- Adatto, I.; Lawrence, C.; Thompson, M.; Zon, L.I. A new system for the rapid collection of large numbers of developmentally staged zebrafish embryos. PLoS ONE 2011, 6, e21715. [Google Scholar] [CrossRef] [PubMed]
- Pardo-Martin, C.; Chang, T.-Y.; Koo, B.K.; Gilleland, C.L.; Wasserman, S.C.; Yanik, M.F. High-throughput in vivo vertebrate screening. Nat. Methods 2010, 7, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Gao, C.; Wang, P.; Zhang, G.-J.; Chen, Z. Fish-on-a-chip: Microfluidics for zebrafish research. Lab Chip 2016, 16, 1106–1125. [Google Scholar] [CrossRef] [PubMed]
- CSEM. Available online: http://www.csem.ch (accessed on 23 May 2017).
- Union Biometrica. Available online: http://www.unionbio.com (accessed on 23 May 2017).

| Zebrafish Drug Screening Study | Number of Compounds Screened | Number of Reported Hits | Reference |
|---|---|---|---|
| Bruni et al. (2016) | 24,760 | Top 100 | [69] |
| Dinday et al. (2015) | 1000 | 4 | [70] |
| Gallardo et al. (2015) | 2960 | 165 | [71] |
| Li et al. (2015) | 3120 | 4 | [72] |
| Nath et al. (2015) | 13,120 | 1 | [73] |
| Liu et al. (2014) | 3000 | 8 | [74] |
| Jin et al. (2013) | 1200 | 6 | [75] |
| Kokel et al. (2013) | 10,000 | 1 Pursued | [76] |
| Nath et al. (2013) | 3120 | 4 | [77] |
| Baxendale et al. (2012) | 2000 | 46 | [78] |
| Kokel et al. (2010) | 14,000 | 1627 | [68] |
| Rihel et al. (2010) | 5648 | 547 | [67] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganzen, L.; Venkatraman, P.; Pang, C.P.; Leung, Y.F.; Zhang, M. Utilizing Zebrafish Visual Behaviors in Drug Screening for Retinal Degeneration. Int. J. Mol. Sci. 2017, 18, 1185. https://doi.org/10.3390/ijms18061185
Ganzen L, Venkatraman P, Pang CP, Leung YF, Zhang M. Utilizing Zebrafish Visual Behaviors in Drug Screening for Retinal Degeneration. International Journal of Molecular Sciences. 2017; 18(6):1185. https://doi.org/10.3390/ijms18061185
Chicago/Turabian StyleGanzen, Logan, Prahatha Venkatraman, Chi Pui Pang, Yuk Fai Leung, and Mingzhi Zhang. 2017. "Utilizing Zebrafish Visual Behaviors in Drug Screening for Retinal Degeneration" International Journal of Molecular Sciences 18, no. 6: 1185. https://doi.org/10.3390/ijms18061185






