The Instrumentation of a Microfluidic Analyzer Enabling the Characterization of the Specific Membrane Capacitance, Cytoplasm Conductivity, and Instantaneous Young’s Modulus of Single Cells
Abstract
:1. Introduction
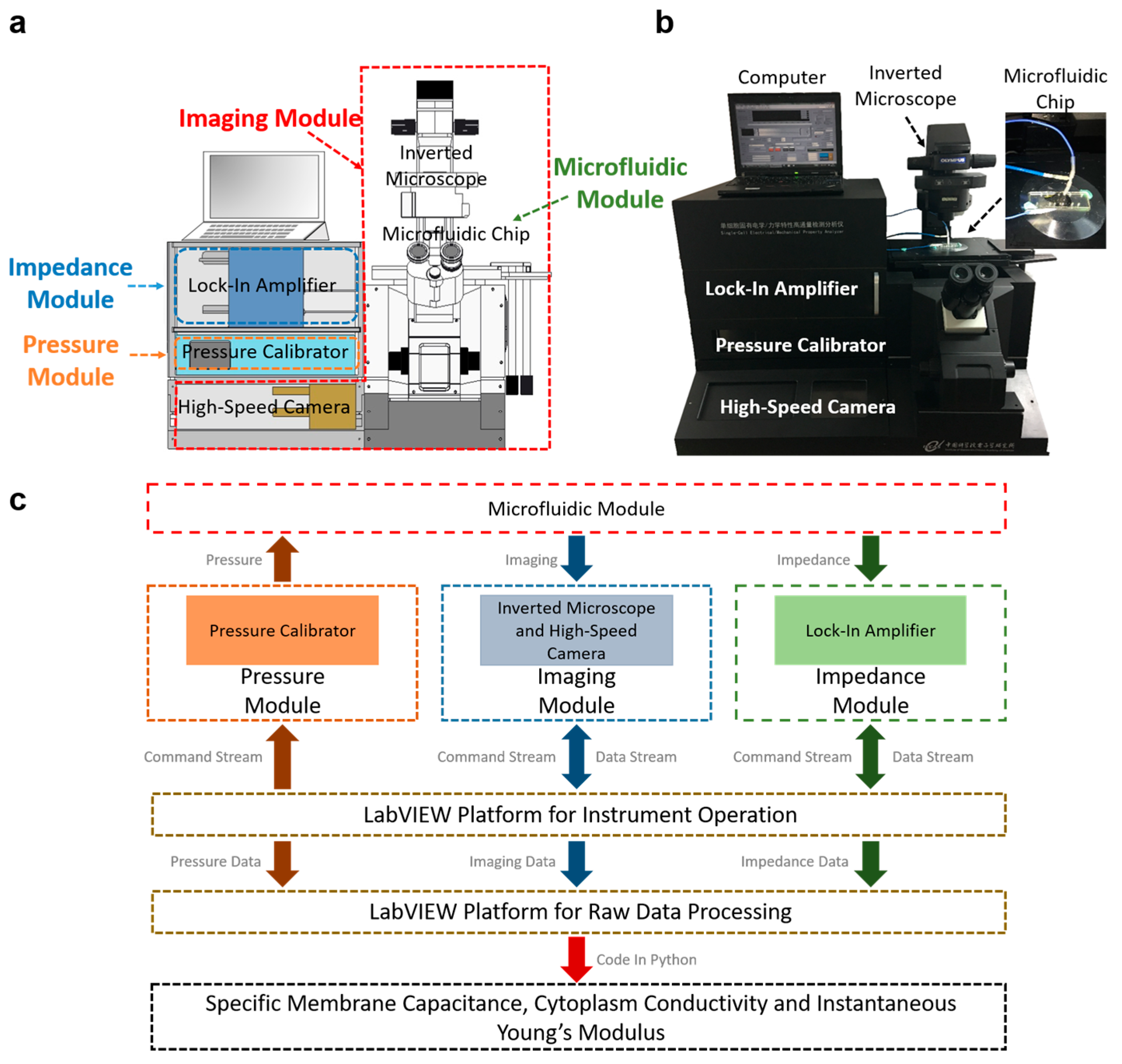
2. Schematics and Working Mechanism
3. Functionalities of Individual Modules
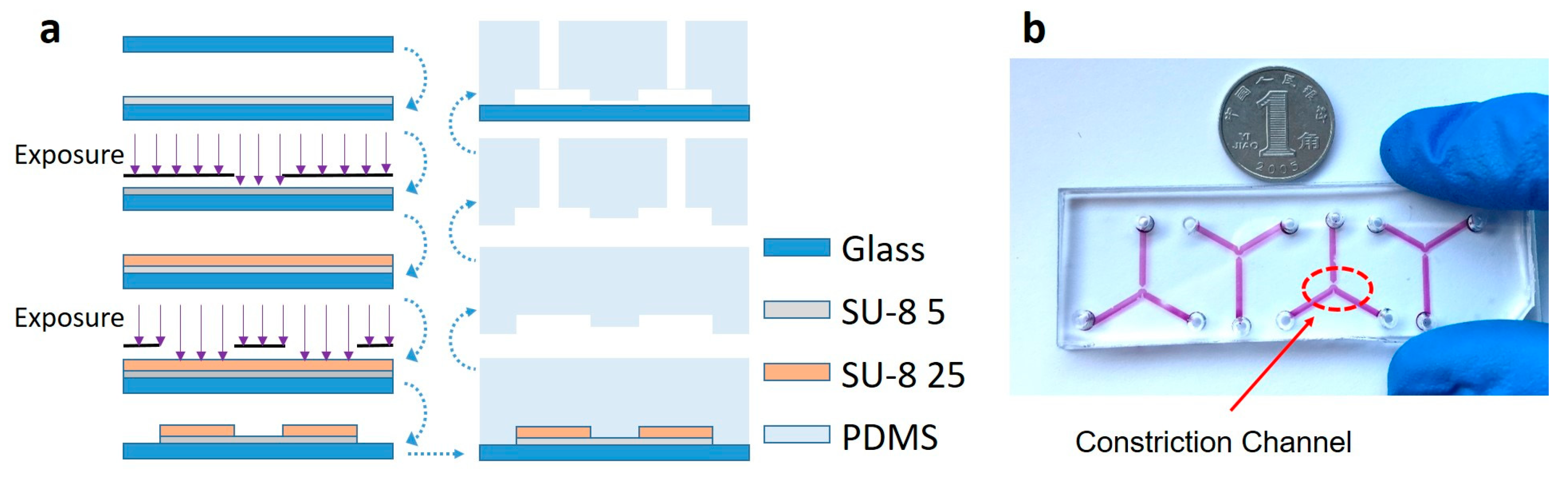
3.1. Microfluidic Module
3.2. Instrument Operation
3.3. Data Processing
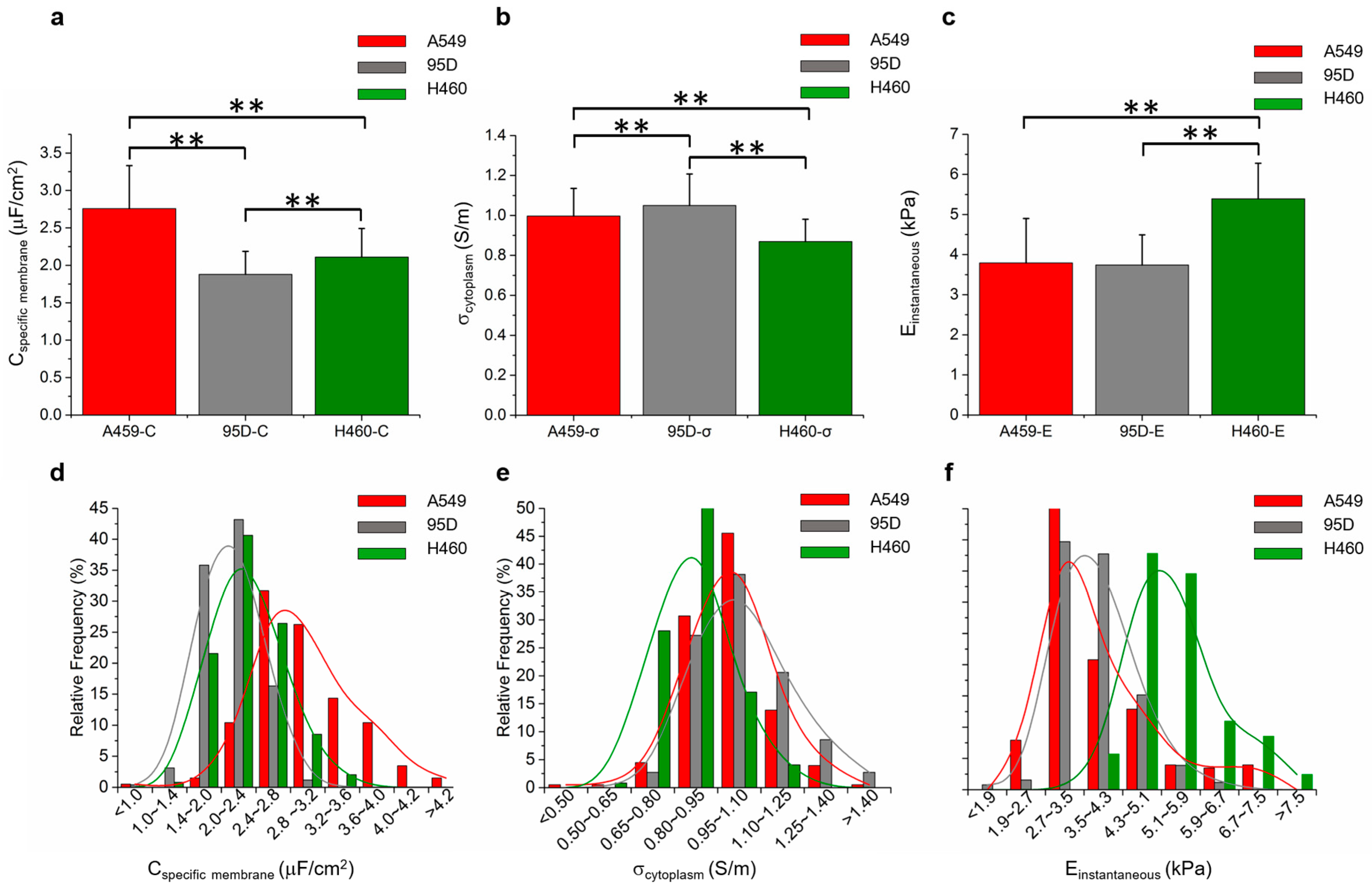
4. Demonstration
5. Conclusions and Future Work
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ethier, C.R.; Simmons, C.A. Introductory Biomechanics: From Cells to Organisms; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Plonsey, R.; Barr, R.C. Bioelectricity: A Quantitative Approach, 3rd ed.; Springer: New York, NY, USA, 2007. [Google Scholar]
- Lee, G.Y.H.; Lim, C.T. Biomechanics approaches to studying human diseases. Trends Biotechnol. 2007, 25, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Di Carlo, D. A mechanical biomarker of cell state in medicine. J. Lab. Autom. 2012, 17, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Xie, X.; Duan, Y.; Wang, L.; Cheng, Z.; Cheng, J. A review of impedance measurements of whole cells. Biosens. Bioelectron. 2016, 77, 824–836. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.; Kondev, J.; Theriot, J.; Garcia, H. Physical Biology of the Cell; Garland Science: New York, NY, USA, 2012. [Google Scholar]
- Arnold, W.M.; Zimmermann, U. Rotating-field-induced rotation and measurement of the membrane capacitance of single mesophyll cells of avena sativa. Zeitschrift Für Naturforschung C 1982, 37, 908–915. [Google Scholar]
- De Gasperis, G.; Wang, X.B.; Yang, J.; Becker, F.F.; Gascoyne, P.R.C. Automated electrorotation: Dielectric characterization of living cells by real-time motion estimation. Meas. Sci. Technol. 1998, 9, 518–529. [Google Scholar] [CrossRef]
- Budde, A.; Grümmer, G.; Knippel, E. Electrorotation of cells and particles: An automated instrumentation. Instrum. Sci. Technol. 1999, 27, 59–66. [Google Scholar] [CrossRef]
- Ino, K.; Ishida, A.; Inoue, K.Y.; Suzuki, M.; Koide, M.; Yasukawa, T.; Shiku, H.; Matsue, T. Electrorotation chip consisting of three-dimensional interdigitated array electrodes. Sens. Actuators B Chem. 2011, 153, 468–473. [Google Scholar] [CrossRef]
- Rohani, A.; Varhue, W.; Su, Y.H.; Swami, N.S. Electrical tweezer for highly parallelized electrorotation measurements over a wide frequency bandwidth. Electrophoresis 2014, 35, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Neher, E.; Marty, A. Discrete changes of cell membrane capacitance observed under conditions of enhanced secretion in bovine adrenal chromaffin cells. Proc. Natl. Acad. Sci. USA 1982, 79, 6712–6716. [Google Scholar] [CrossRef] [PubMed]
- Rituper, B. High-resolution membrane capacitance measurements for the study of exocytosis and endocytosis. Nat. Protoc. 2013, 8, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Qu, A.; Luo, J. Error analysis of cm measurement under the whole-cell patch-clamp recording. J. Neurosci. Methods 2010, 185, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Neef, A.; Heinemann, C.; Moser, T. Measurements of membrane patch capacitance using a software-based lock-in system. Pflüg. Arch. Eur. J. Physiol. 2007, 454, 335–344. [Google Scholar] [CrossRef] [PubMed]
- O’Shaughnessy, T.J.; Kim, Y.I. A computer-based system for the measurement of membrane capacitance to monitor exocytosis in secretory cells. J. Neurosci. Methods 1995, 57, 1–8. [Google Scholar] [CrossRef]
- Cho, Y.; Kim, H.S.; Frazier, A.B.; Chen, Z.G.; Shin, D.M.; Han, A. Whole-cell impedance analysis for highly and poorly metastatic cancer cells. J. Microelectromech. Syst. 2009, 18, 808–817. [Google Scholar]
- Ghenim, L.; Kaji, H.; Hoshino, Y.; Ishibashi, T.; Haguet, V.; Gidrol, X.; Nishizawa, M. Monitoring impedance changes associated with motility and mitosis of a single cell. Lab Chip 2010, 10, 2546–2550. [Google Scholar] [CrossRef] [PubMed]
- Labeed, F.H.; Coley, H.M.; Hughes, M.P. Differences in the biophysical properties of membrane and cytoplasm of apoptotic cells revealed using dielectrophoresis. Biochim. Biophys. Acta 2006, 1760, 922–929. [Google Scholar] [CrossRef] [PubMed]
- Coley, H.M.; Labeed, F.H.; Thomas, H.; Hughes, M.P. Biophysical characterization of mdr breast cancer cell lines reveals the cytoplasm is critical in determining drug sensitivity. Biochim. Biophys. Acta 2007, 1770, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.A. New membrane concept applied to the analysis of fluid shear- and micropipette-deformed red blood cells. Biophys. J. 1973, 13, 941–954. [Google Scholar] [CrossRef]
- Zhou, E.H.; Lim, C.T.; Quek, S.T. Finite element simulation of the micropipette aspiration of a living cell undergoing large viscoelastic deformation. Mech. Adv. Mater. Struct. 2005, 12, 501–512. [Google Scholar] [CrossRef]
- He, J.H.; Xu, W.; Zhu, L. Analytical model for extracting mechanical properties of a single cell in a tapered micropipette. Appl. Phys. Lett. 2007, 90, 023901. [Google Scholar] [CrossRef]
- Zhao, Q.; Wu, M.; Cui, M.; Qin, Y.; Yu, J.; Sun, M.; Zhao, X.; Feng, X. A novel pneumatic micropipette aspiration method using a balance pressure model. Rev. Sci. Instrum. 2013, 84, 123703. [Google Scholar] [CrossRef] [PubMed]
- Shojaei-Baghini, E.; Zheng, Y.; Sun, Y. Automated micropipette aspiration of single cells. Ann. Biomed. Eng. 2013, 41, 1208–1216. [Google Scholar] [CrossRef] [PubMed]
- Hoh, J.H.; Schoenenberger, C.A. Surface morphology and mechanical properties of mdck monolayers by atomic force microscopy. J. Cell Sci. 1994, 107 Pt 5, 1105–1114. [Google Scholar] [PubMed]
- Takahashi, R.; Okajima, T. Mapping power-law rheology of living cells using multi-frequency force modulation atomic force microscopy. Appl. Phys. Lett. 2015, 107, 1564. [Google Scholar] [CrossRef]
- Lekka, M.; Pogoda, K.; Gostek, J.; Klymenko, O.; Prauzner-Bechcicki, S.; Wiltowska-Zuber, J.; Jaczewska, J.; Lekki, J.; Stachura, Z. Cancer cell recognition-mechanical phenotype. Micron 2012, 43, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Braunsmann, C.; Seifert, J.; Rheinlaender, J.; Schaffer, T.E. High-speed force mapping on living cells with a small cantilever atomic force microscope. Rev. Sci. Instrum. 2014, 85, 073703. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Nguyen, J.; Wei, Y.; Sun, Y. Recent advances in microfluidic techniques for single-cell biophysical characterization. Lab Chip 2013, 13, 2464–2483. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, Y.; Tan, Q.; Zhang, Y.L.; Li, J.; Geddie, W.R.; Jewett, M.A.; Sun, Y. A microfluidic device for simultaneous electrical and mechanical measurements on single cells. Biomicrofluidics 2011, 5, 14113. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zheng, Y.; Tan, Q.; Shojaei-Baghini, E.; Zhang, Y.L.; Li, J.; Prasad, P.; You, L.; Wu, X.Y.; Sun, Y. Classification of cell types using a microfluidic device for mechanical and electrical measurement on single cells. Lab Chip 2011, 11, 3174–3181. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Shojaei-Baghini, E.; Azad, A.; Wang, C.; Sun, Y. High-throughput biophysical measurement of human red blood cells. Lab Chip 2012, 12, 2560–2567. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, D.; Luo, Y.; Chen, F.; Zhao, X.; Jiang, M.; Yue, W.; Long, R.; Wang, J.; Chen, J. Simultaneous characterization of instantaneous young’s modulus and specific membrane capacitance of single cells using a microfluidic system. Sensors 2015, 15, 2763–2773. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, X.T.; Chen, D.Y.; Luo, Y.N.; Jiang, M.; Wei, C.; Long, R.; Yue, W.T.; Wang, J.B.; Chen, J. Tumor cell characterization and classification based on cellular specific membrane capacitance and cytoplasm conductivity. Biosens. Bioelectron. 2014, 57, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.N.; Chen, D.Y.; Zhao, Y.; Wei, C.; Zhao, X.T.; Yue, W.T.; Long, R.; Wang, J.B.; Chen, J. A constriction channel based microfluidic system enabling continuous characterization of cellular instantaneous young’s modulus. Sens. Actuators B Chem. 2014, 202, 1183–1189. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, K.; Zhao, Y.; Chen, D.; Huang, C.; Fan, B.; Long, R.; Hsieh, C.-H.; Wang, J.; Wu, M.-H.; Chen, J. The Instrumentation of a Microfluidic Analyzer Enabling the Characterization of the Specific Membrane Capacitance, Cytoplasm Conductivity, and Instantaneous Young’s Modulus of Single Cells. Int. J. Mol. Sci. 2017, 18, 1158. https://doi.org/10.3390/ijms18061158
Wang K, Zhao Y, Chen D, Huang C, Fan B, Long R, Hsieh C-H, Wang J, Wu M-H, Chen J. The Instrumentation of a Microfluidic Analyzer Enabling the Characterization of the Specific Membrane Capacitance, Cytoplasm Conductivity, and Instantaneous Young’s Modulus of Single Cells. International Journal of Molecular Sciences. 2017; 18(6):1158. https://doi.org/10.3390/ijms18061158
Chicago/Turabian StyleWang, Ke, Yang Zhao, Deyong Chen, Chengjun Huang, Beiyuan Fan, Rong Long, Chia-Hsun Hsieh, Junbo Wang, Min-Hsien Wu, and Jian Chen. 2017. "The Instrumentation of a Microfluidic Analyzer Enabling the Characterization of the Specific Membrane Capacitance, Cytoplasm Conductivity, and Instantaneous Young’s Modulus of Single Cells" International Journal of Molecular Sciences 18, no. 6: 1158. https://doi.org/10.3390/ijms18061158






