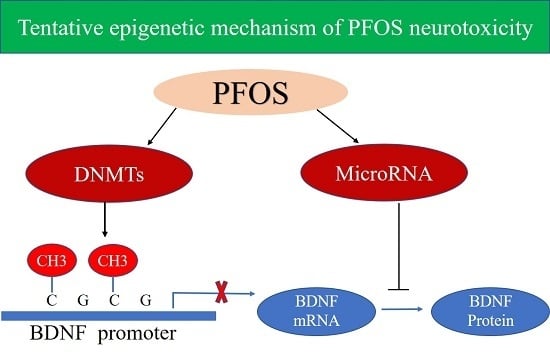
Brain-Derived Neurotrophic Factor Mediated Perfluorooctane Sulfonate Induced-Neurotoxicity via Epigenetics Regulation in SK-N-SH Cells
Abstract
:1. Introduction
2. Results
2.1. Morphology Analysis of SK-N-SH Cell after PFOS Exposure
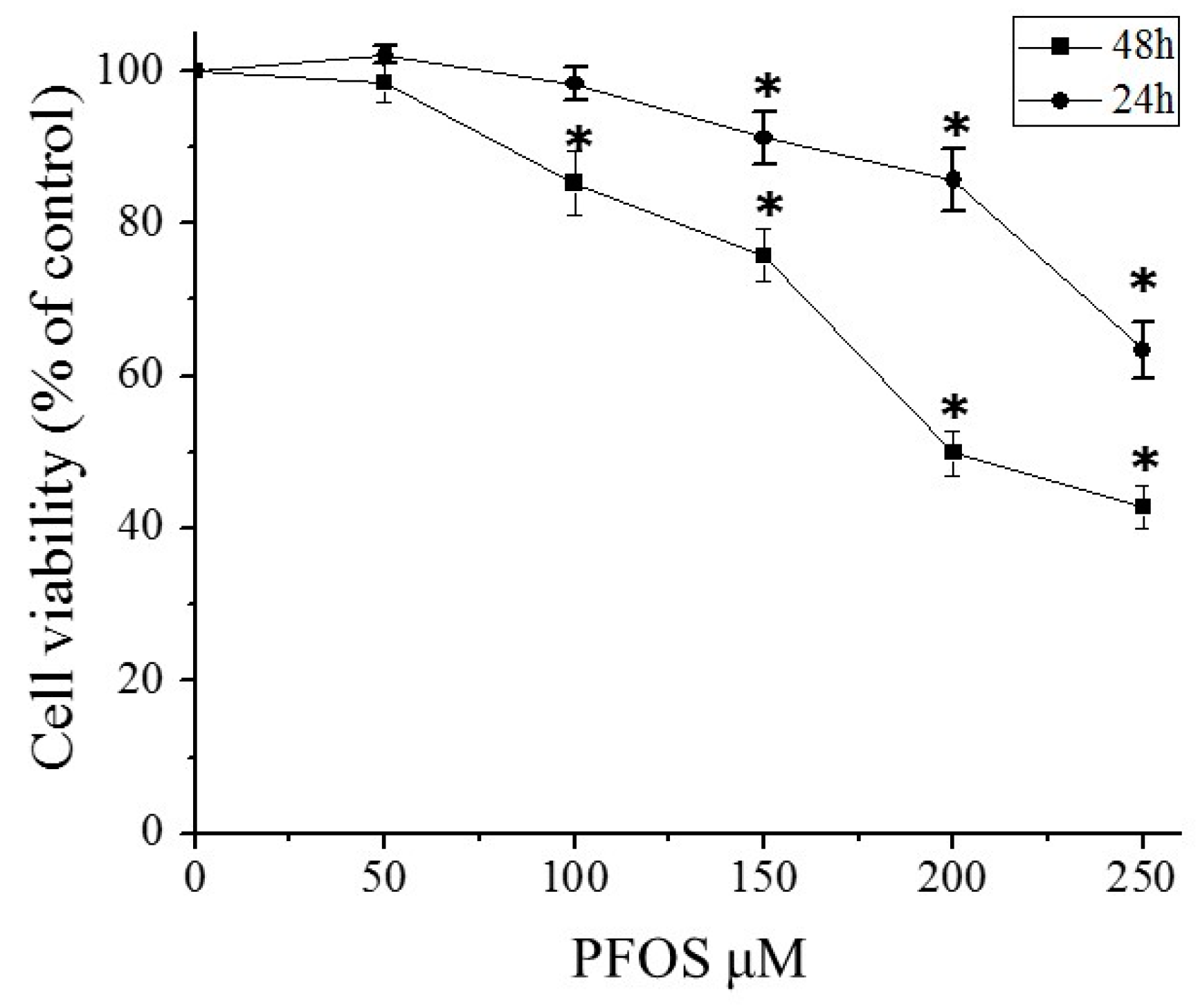
2.2. PFOS Inhibited Cell Growth in SK-N-SH Cells
2.3. PFOS Reduced the Expression of BDNF
2.4. The Effects of PFOS on the BDNF Promoter Methylation in SK-N-SH Cells
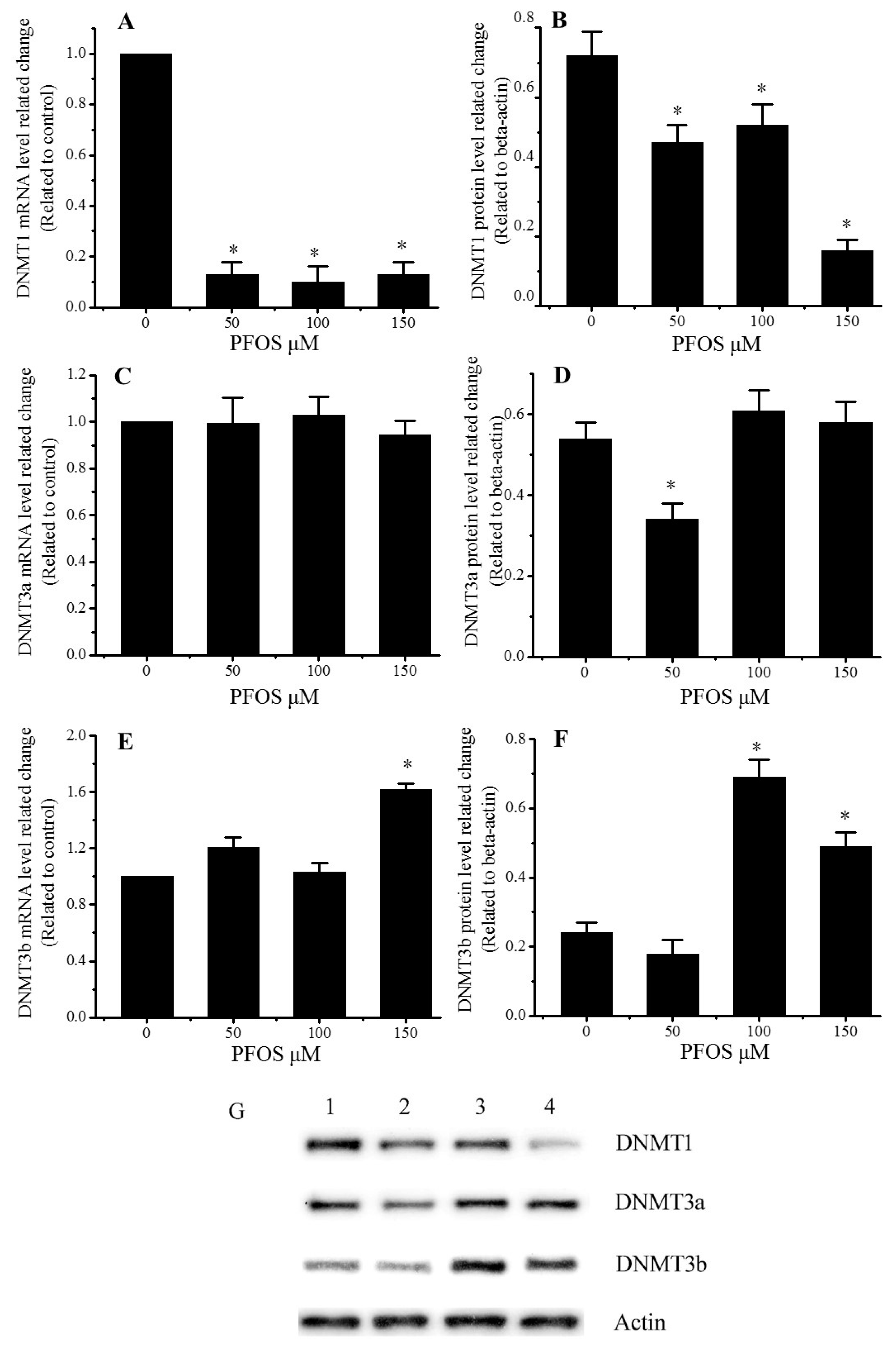
2.5. Effect of PFOS on the Expression of DNMT1, DNMT3a, and DNMT3b in SK-N-SH Cells
2.6. PFOS Increases the Expression of MicroRNA-16, MicroRNA-22, and MicroRNA-30a-5p
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Cell Culture and PFOS Treatment
4.3. Morphological Study and Cell Viability Assay (MTT)
4.4. RNA Extraction and cDNA Synthesis
4.5. Real-Time Quantitative PCR (Q-PCR)
4.6. Enzyme Linked Immunosorbent Assay (ELISA)
4.7. Western Blot Assays
4.8. Determination of the Status of BDNF Promoter Methylation
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Giesy, J.P.; Kannan, K. Perfluorochemical surfactants in the environment. Environ. Sci. Technol. 2002, 36, 146A–152A. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Li, J.; Li, D.; Yang, Y.; He, D. Chronic exposure to perfluorooctane sulfonate induces behavior defects and neurotoxicity through oxidative damages, in vivo and in vitro. PLoS ONE 2014, 9, e113453. [Google Scholar] [CrossRef] [PubMed]
- Bhavsar, S.P.; Fowler, C.; Day, S.; Petro, S.; Gandhi, N.; Gewurtz, S.B.; Hao, C.; Zhao, X.; Drouillard, K.G.; Morse, D. High levels, partitioning and fish consumption based water guidelines of perfluoroalkyl acids downstream of a former firefighting training facility in canada. Environ. Int. 2016, 94, 415–423. [Google Scholar] [CrossRef] [PubMed]
- Miralles-Marco, A.; Harrad, S. Perfluorooctane sulfonate: A review of human exposure, biomonitoring and the environmental forensics utility of its chirality and isomer distribution. Environ. Int. 2015, 77, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Saikat, S.; Kreis, I.; Davies, B.; Bridgman, S.; Kamanyire, R. The impact of pfos on health in the general population: A review. Environ. Sci. Process. Impacts 2013, 15, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Croce, N.; Gelfo, F.; Ciotti, M.T.; Federici, G.; Caltagirone, C.; Bernardini, S.; Angelucci, F. Npy modulates mir-30a-5p and bdnf in opposite direction in an in vitro model of Alzheimer disease: A possible role in neuroprotection? Mol. Cell Biochem. 2013, 376, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Daito, T.; Sasaki, Y.; Chung, Y.H.; Xing, X.; Pondugula, S.; Swamidass, S.J.; Wang, T.; Kim, A.H.; Yano, H. Inhibition of DNA methyltransferases blocks mutant huntingtin-induced neurotoxicity. Sci. Rep. 2016, 6, 31022. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Liu, X.L.; Li, J.; Mu, R.H.; Liu, Q.; Yi, L.T.; Geng, D. Macranthol promotes hippocampal neuronal proliferation in mice via BDNF-TrkB-PI3K/Akt signaling pathway. Eur. J. Pharmacol. 2015, 762, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, T.B.; Pinton, S.; da Rocha, J.T.; Gai, B.M.; Nogueira, C.W. Involvement of BDNF/TrkB signaling in the effect of diphenyl diselenide on motor function in a parkinson’s disease rat model. Eur. J. Pharmacol. 2016, 795, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, W.; Zhang, Q.; Zhao, H.; Quan, X. Effects of developmental perfluorooctane sulfonate exposure on spatial learning and memory ability of rats and mechanism associated with synaptic plasticity. Food Chem. Toxicol. 2015, 76, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, Q.Z.; Wu, C.Q.; Pan, X.Y.; Wang, J.; Tan, Y.; Shan, X.Y.; Zeng, H.C. PFOS disturbs BDNF-ERK-CREB signalling in association with increased microRNA-22 in SH-SY5Y cells. BioMed Res. Int. 2015, 2015, 302653. [Google Scholar] [CrossRef] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Numakawa, T.; Richards, M.; Adachi, N.; Kishi, S.; Kunugi, H.; Hashido, K. MicroRNA function and neurotrophin bdnf. Neurochem. Int. 2011, 59, 551–558. [Google Scholar] [CrossRef] [PubMed]
- You, H.J.; Park, J.H.; Pareja-Galeano, H.; Lucia, A.; Shin, J.I. Targeting microRNAs involved in the BDNF signaling impairment in neurodegenerative diseases. Neuromol. Med. 2016, 18, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Huang, H.S.; Grigorenko, A.; Rogaev, E.; Akbarian, S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 2008, 17, 3030–3042. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Chen, L.; Ding, J.; Zhang, J.; Fan, Z.; Yang, C.; Yu, Q.; Yang, J. Cardioprotective effect of miRNA-22 on hypoxia/reoxygenation induced cardiomyocyte injury in neonatal rats. Gene 2016, 579, 17–22. [Google Scholar] [CrossRef] [PubMed]
- Muinos-Gimeno, M.; Espinosa-Parrilla, Y.; Guidi, M.; Kagerbauer, B.; Sipila, T.; Maron, E.; Pettai, K.; Kananen, L.; Navines, R.; Martin-Santos, R.; et al. Human microRNAs miR-22, miR-138–2, miR-148a, and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol. Psychiatr. 2011, 69, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Zhang, Y. DNA methylation in mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, W.; Liu, S.; Zong, S.; Wang, W.; Ren, J.; Li, Q.; Hou, F.; Shi, Q. DNMT1, DNMT3A and DNMT3B polymorphisms associated with gastric cancer risk: A systematic review and meta-analysis. EBioMedicine 2016, 13, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Preston, R.; Goldman, L.R.; Brebi-Mieville, P.; Ili-Gangas, C.; LeBron, C.; Witter, F.R.; Apelberg, B.J.; Hernández-Roystacher, M.; Jaffe, A.; Halden, R.U.; et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics 2014, 5, 539–546. [Google Scholar] [CrossRef]
- Wan, Y.J.; Li, Y.Y.; Xia, W.; Chen, J.; Lv, Z.Q.; Zeng, H.C.; Zhang, L.; Yang, W.J.; Chen, T.; Lin, Y.; et al. Alterations in tumor biomarker GSTP gene methylation patterns induced by prenatal exposure to PFOS. Toxicology 2010, 274, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ramos, J.C.; Gutierrez, A.G.; Vazquez-Aguirre, A.; Toledano-Magana, Y.; Alonso-Saenz, A.L.; Gomez-Vidales, V.; Flores-Alamo, M.; Mejia, C.; Ruiz-Azuara, L. The mitochondrial apoptotic pathway is induced by Cu(II) antineoplastic compounds (Casiopeinas®) in SK-N-SH neuroblastoma cells after short exposure times. Biometals 2017, 30, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Yogarajah, T.; Ong, K.C.; Perera, D.; Wong, K.T. Enterovirus a71 and coxsackievirus a16 show different replication kinetics in human neuronal and non-neuronal cell lines. Arch. Virol. 2016, 162, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Johansson, N.; Fredriksson, A.; Eriksson, P. Neonatal exposure to perfluorooctane sulfonate (PFOS) and perfluorooctanoic acid (PFOA) causes neurobehavioural defects in adult mice. Neurotoxicology 2008, 29, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Johansson, N.; Eriksson, P.; Viberg, H. Neonatal exposure to PFOS and pfoa in mice results in changes in proteins which are important for neuronal growth and synaptogenesis in the developing brain. Toxicol. Sci. 2009, 108, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.C.; Li, Y.Y.; Zhang, L.; Wang, Y.J.; Chen, J.; Xia, W.; Lin, Y.; Wei, J.; Lv, Z.Q.; Li, M.; et al. Prenatal exposure to perfluorooctanesulfonate in rat resulted in long-lasting changes of expression of synapsins and synaptophysin. Synapse 2011, 65, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Oh, H.; Lewis, D.A.; Sibille, E. The role of bdnf in age-dependent changes of excitatory and inhibitory synaptic markers in the human prefrontal cortex. Neuropsychopharmacology 2016, 41, 3080–3091. [Google Scholar] [CrossRef] [PubMed]
- Bramham, C.R.; Messaoudi, E. BDNF function in adult synaptic plasticity: The synaptic consolidation hypothesis. Prog. Neurobiol. 2005, 76, 99–125. [Google Scholar] [CrossRef] [PubMed]
- Autry, A.E.; Monteggia, L.M. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol. Rev. 2012, 64, 238–258. [Google Scholar] [CrossRef] [PubMed]
- Karpova, N.N. Role of BDNF epigenetics in activity-dependent neuronal plasticity. Neuropharmacology 2014, 76 Pt C, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Muller, S. In silico analysis of regulatory networks underlines the role of miR-10b-5p and its target BDNF in huntington’s disease. Transl. Neurodegener. 2014, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.X.; Yang, J.; Wang, P.Y.; Li, Y.J.; Xie, S.Y.; Sun, R.P. Cisplatin regulates SH-SY5Y cell growth through downregulation of bdnf via miR-16. Oncol. Rep. 2013, 30, 2343–2349. [Google Scholar] [PubMed]
- Chen, Y.L.; Shen, C.K. Modulation of mGluR-dependent MAP1B translation and AMPA receptor endocytosis by microRNA miR-146a-5p. J. Neurosci. 2013, 33, 9013–9020. [Google Scholar] [CrossRef] [PubMed]
- Toledo-Rodriguez, M.; Lotfipour, S.; Leonard, G.; Perron, M.; Richer, L.; Veillette, S.; Pausova, Z.; Paus, T. Maternal smoking during pregnancy is associated with epigenetic modifications of the brain-derived neurotrophic factor-6 exon in adolescent offspring. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010, 153B, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Roth, T.L.; Lubin, F.D.; Funk, A.J.; Sweatt, J.D. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol. Psychiatr. 2009, 65, 760–769. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.S.; Jia, M.; Sun, J.; Sun, X.R.; Zhang, H.; Ji, M.H.; Yang, J.J.; Wang, Z.Y. Hypermethylation of hippocampal synaptic plasticity-related genes is involved in neonatal sevoflurane exposure-induced cognitive impairments in rats. Neurotox. Res. 2016, 29, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Anier, K.; Malinovskaja, K.; Pruus, K.; Aonurm-Helm, A.; Zharkovsky, A.; Kalda, A. Maternal separation is associated with DNA methylation and behavioural changes in adult rats. Eur. Neuropsychopharmacol. 2014, 24, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; Wang, Y.; Ju, L.H.; Chen, M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiol. Learn. Mem. 2012, 97, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Bestor, T.H.; Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992, 69, 915–926. [Google Scholar] [CrossRef]
- Okano, M.; Xie, S.; Li, E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19, 219–220. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]




| Gene Name | Primer | Product (bp) |
|---|---|---|
| BDNF | Forward: AGCTGAGCGTGTGTGACAGTATTAG | 129 |
| Reverse: ATTGCTTCAGTTGGCCTTTTGATAC | ||
| DNMT1 | Forward: CGACTACATCAAAGGCAGCA | 161 |
| Reverse: TGGACTTGTGGGTGTTCTCA | ||
| DNMT3a | Forward: GCATTGTGTCTTGGTGGATG | 191 |
| Reverse: GACCTCGTAGATGGCTTTGC | ||
| DNMT3b | Forward: AGGATGGGAAGGAGTTTGGA | 114 |
| Reverse: ACATAGCCTGTCGCTTGGAG | ||
| Actin | Forward: TGAAGGTGACAGCAGTCGGTT | 126 |
| Reverse: ACTTCCTGTAACAACGCATCTCATA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.-X.; He, Q.-Z.; Li, W.; Long, D.-X.; Pan, X.-Y.; Chen, C.; Zeng, H.-C. Brain-Derived Neurotrophic Factor Mediated Perfluorooctane Sulfonate Induced-Neurotoxicity via Epigenetics Regulation in SK-N-SH Cells. Int. J. Mol. Sci. 2017, 18, 893. https://doi.org/10.3390/ijms18040893
Guo X-X, He Q-Z, Li W, Long D-X, Pan X-Y, Chen C, Zeng H-C. Brain-Derived Neurotrophic Factor Mediated Perfluorooctane Sulfonate Induced-Neurotoxicity via Epigenetics Regulation in SK-N-SH Cells. International Journal of Molecular Sciences. 2017; 18(4):893. https://doi.org/10.3390/ijms18040893
Chicago/Turabian StyleGuo, Xin-Xin, Qing-Zhi He, Wu Li, Ding-Xin Long, Xiao-Yuan Pan, Cong Chen, and Huai-Cai Zeng. 2017. "Brain-Derived Neurotrophic Factor Mediated Perfluorooctane Sulfonate Induced-Neurotoxicity via Epigenetics Regulation in SK-N-SH Cells" International Journal of Molecular Sciences 18, no. 4: 893. https://doi.org/10.3390/ijms18040893