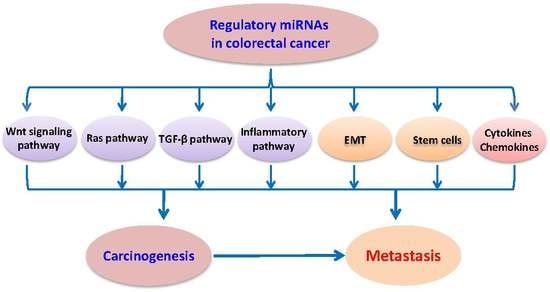
Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis
Abstract
:1. Introduction
2. Clinical Significance of miRNAs in Colorectal Cancer
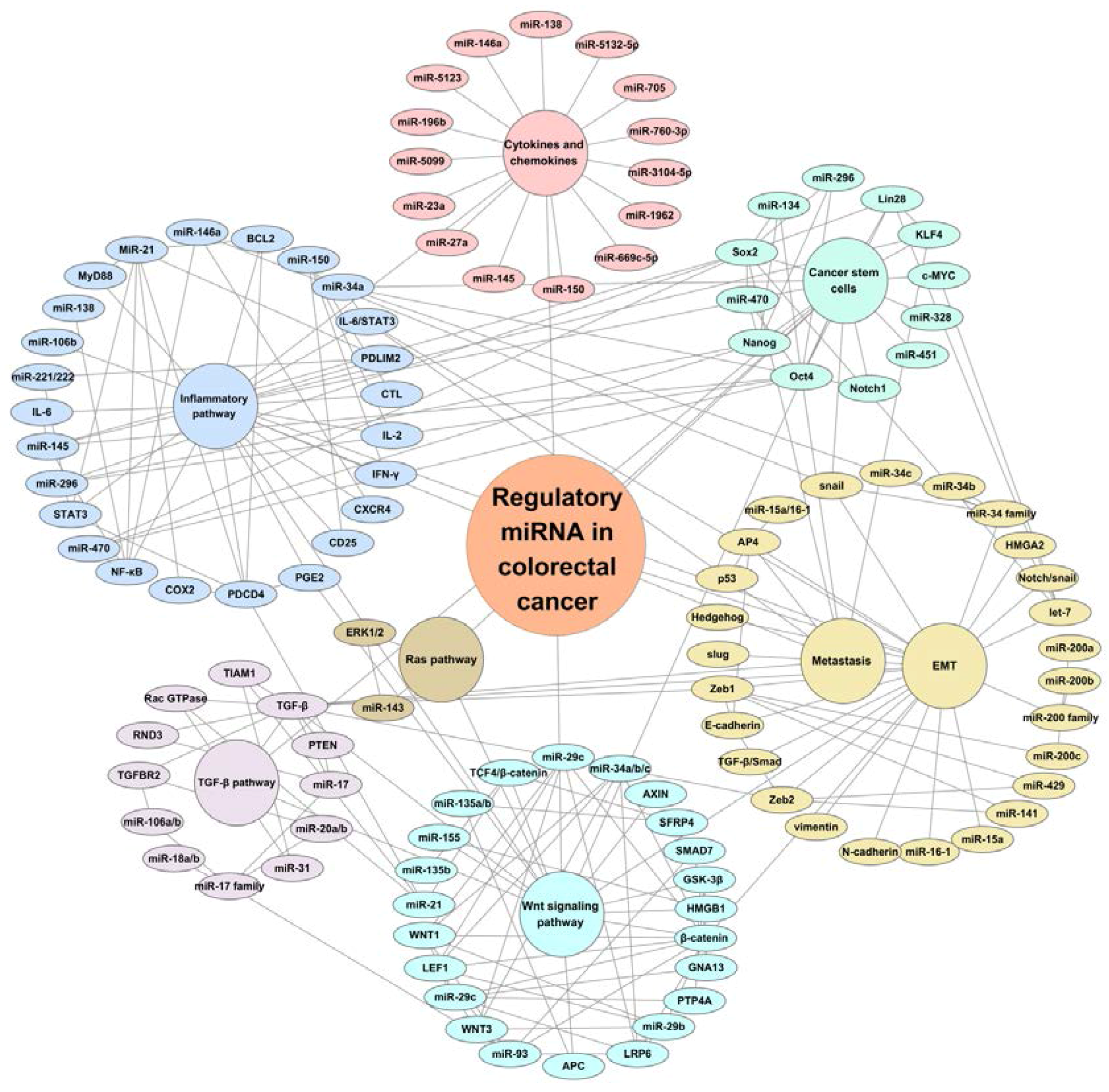
3. Regulatory miRNAs in Oncogenic Signaling Pathways
3.1. miRNAs and Wnt Signaling
3.2. miRNAs and RAS Pathway
3.3. miRNAs and TGF-β Pathway
3.4. miRNAs and Inflammatory Pathway
4. Regulatory miRNAs in Cancer Stem Cells, Epithelial-Mesenchymal Transition (EMT), and Metastasis
4.1. miRNAs and Cancer Stem Cells
4.2. miRNAs and EMT and Metastasis
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| miRNA | microRNA |
| CRC | colorectal cancer |
| CSC | cancer stem cells |
| EMT | epithelial-mesenchymal transition |
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Punt, C.J.; Koopman, M.; Vermeulen, L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat. Rev. Clin. Oncol. 2017, 14, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Raskov, H.; Pommergaard, H.C.; Burcharth, J.; Rosenberg, J. Colorectal carcinogenesis-update and perspectives. World J. Gastroenterol. 2014, 20, 18151–18164. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I. Inflammation and colorectal cancer: Colitis-associated neoplasia. Semin. Immunopathol. 2013, 35, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Brenner, H.; Kloor, M.; Pox, C.P. Colorectal cancer. Lancet 2013, 383, 1490–1502. [Google Scholar] [CrossRef]
- Schetter, A.J.; Harris, C.C. Alterations of microRNAs contribute to colon carcinogenesis. Semin. Oncol. 2011, 38, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.V.; Galbraith, N.J.; Yang, D.; Burton, J.F.; Walker, S.P.; Galandiuk, S. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: A systematic review and meta-analysis. Br. J. Cancer 2017, 116, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Strubberg, A.M.; Madison, B.B. MicroRNAs in the etiology of colorectal cancer: Pathways and clinical implications. Dis. Models Mech. 2017, 10, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Marmol, I.; Sanchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Moridikia, A.; Mirzaei, H.; Sahebkar, A.; Salimian, J. MicroRNAs: Potential Candidates for Diagnosis and Treatment of Colorectal Cancer. J. Cell. Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015. [Google Scholar] [CrossRef] [PubMed]
- Mamoori, A.; Gopalan, V.; Smith, R.A.; Lam, A.K. Modulatory roles of microRNAs in the regulation of different signalling pathways in large bowel cancer stem cells. Biol. Cell 2016, 108, 51–64. [Google Scholar] [CrossRef] [PubMed]
- De Krijger, I.; Mekenkamp, L.J.; Punt, C.J.; Nagtegaal, I.D. MicroRNAs in colorectal cancer metastasis. J. Pathol. 2011, 224, 438–447. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Du, Y.; Liu, X.; Cho, W.C.; Yang, Y. MicroRNAs as Regulator of Signaling Networks in Metastatic Colon Cancer. BioMed Res. Int. 2015, 2015, 823620. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Guo, Y.; Li, Z.; Fang, W.; Yang, Y.; Li, X.; Li, Z.; Xiong, B.; Chen, Z.; Wang, J.; et al. MicroRNA profiling in Muc2 knockout mice of colitis-associated cancer model reveals epigenetic alterations during chronic colitis malignant transformation. PLoS ONE 2014, 9, e99132. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Wang, Z.; Fan, Q.; Wang, R.; Sun, Y. microRNA-27a functions as a tumor suppressor in esophageal squamous cell carcinoma by targeting KRAS. Oncol. Rep. 2014, 31, 280–286. [Google Scholar] [PubMed]
- Scheibner, K.A.; Teaboldt, B.; Hauer, M.C.; Chen, X.; Cherukuri, S.; Guo, Y.; Kelley, S.M.; Liu, Z.; Baer, M.R.; Heimfeld, S.; et al. miR-27a functions as a tumor suppressor in acute leukemia by regulating 14-3-3θ. PLoS ONE 2012, 7, e50895. [Google Scholar] [CrossRef] [PubMed]
- Chintharlapalli, S.; Papineni, S.; Abdelrahim, M.; Abudayyeh, A.; Jutooru, I.; Chadalapaka, G.; Wu, F.; Mertens-Talcott, S.; Vanderlaag, K.; Cho, S.D.; et al. Oncogenic microRNA-27a is a target for anticancer agent methyl 2-cyano-3,11-dioxo-18β-olean-1,12-dien-30-oate in colon cancer cells. Int. J. Cancer 2009, 125, 1965–1974. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Mansoori, B.; Baradaran, B. The role of microRNAs in colorectal cancer. Biomed. Pharmacother. 2016, 84, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Chen, Z.; Guo, Y.; Feng, Y.; Li, Z.; Han, W.; Wang, J.; Zhao, W.; Jiao, Y.; Li, K.; et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and Smad2. PLoS ONE 2014, 9, e105991. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chen, L.; Xu, Y.; Li, R.; Du, X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2010, 400, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wu, J.; Meng, X.; Ying, X.; Zuo, Y.; Liu, R.; Pan, Z.; Kang, T.; Huang, W. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis 2011, 32, 1033–1042. [Google Scholar] [CrossRef] [PubMed]
- Hollis, M.; Nair, K.; Vyas, A.; Chaturvedi, L.S.; Gambhir, S.; Vyas, D. MicroRNAs potential utility in colon cancer: Early detection, prognosis, and chemosensitivity. World J. Gastroenterol. 2015, 21, 8284–8292. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ma, D.; Fesler, A.; Zhai, H.; Leamniramit, A.; Li, W.; Wu, S.; Ju, J. Expression analysis of microRNA as prognostic biomarkers in colorectal cancer. Oncotarget 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X.; Song, W.; Chen, Z.H.; Wei, J.H.; Liao, Y.J.; Lei, J.; Hu, M.; Chen, G.Z.; Liao, B.; Lu, J.; et al. Prognostic and predictive value of a microRNA signature in stage II colon cancer: A microRNA expression analysis. Lancet 2013, 14, 1295–1306. [Google Scholar] [CrossRef]
- Pichler, M.; Winter, E.; Stotz, M.; Eberhard, K.; Samonigg, H.; Lax, S.; Hoefler, G. Down-regulation of KRAS-interacting miRNA-143 predicts poor prognosis but not response to EGFR-targeted agents in colorectal cancer. Br. J. Cancer 2012, 106, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Aoki, K.; Taketo, M.M. Adenomatous polyposis coli (APC): A multi-functional tumor suppressor gene. J. Cell Sci. 2007, 120 Pt 19, 3327–3335. [Google Scholar] [CrossRef] [PubMed]
- Nathke, I. Cytoskeleton out of the cupboard: Colon cancer and cytoskeletal changes induced by loss of APC. Nat. Rev. Cancer 2006, 6, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Segditsas, S.; Tomlinson, I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene 2006, 25, 7531–7537. [Google Scholar] [CrossRef] [PubMed]
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Oude Vrielink, J.A.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802. [Google Scholar] [CrossRef] [PubMed]
- Kawano, Y.; Kypta, R. Secreted antagonists of the Wnt signalling pathway. J. Cell Sci. 2003, 116 Pt 13, 2627–2634. [Google Scholar] [CrossRef] [PubMed]
- Valeri, N.; Braconi, C.; Gasparini, P.; Murgia, C.; Lampis, A.; Paulus-Hock, V.; Hart, J.R.; Ueno, L.; Grivennikov, S.I.; Lovat, F.; et al. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014, 25, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.L.; Wu, D.W.; Huang, C.C.; He, T.Y.; Chou, M.C.; Sheu, G.T.; Lee, H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis 2014, 35, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Yue, X.; Han, L.; Shi, Z.; Yang, Y.; Pu, P.; Yao, Z.; Kang, C. Genome-wide identification of TCF7L2/TCF4 target miRNAs reveals a role for miR-21 in Wnt-driven epithelial cancer. Int. J. Oncol. 2012, 40, 519–526. [Google Scholar] [PubMed]
- Wan, J.; Xia, L.; Xu, W.; Lu, N. Expression and Function of miR-155 in Diseases of the Gastrointestinal Tract. Int. J. Mol. Sci. 2016, 17, 709. [Google Scholar] [CrossRef] [PubMed]
- Itou, J.; Taniguchi, N.; Oishi, I.; Kawakami, H.; Lotz, M.; Kawakami, Y. HMGB factors are required for posterior digit development through integrating signaling pathway activities. Dev. Dyn. 2011, 240, 1151–1162. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, H.S.; Kim, N.G.; Lee, I.; Choi, H.S.; Li, X.Y.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Na, J.M.; et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 2011, 4, ra71. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, M.; Rao, S.R.; Thacker, P.; Chatterjee, S.; Karunagaran, D. miR-29b downregulates canonical Wnt signaling by suppressing coactivators of β-catenin in human colorectal cancer cells. J. Cell. Biochem. 2014, 115, 1974–1984. [Google Scholar] [PubMed]
- Zhang, J.X.; Mai, S.J.; Huang, X.X.; Wang, F.W.; Liao, Y.J.; Lin, M.C.; Kung, H.F.; Zeng, Y.X.; Xie, D. miR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann. Oncol. 2014, 25, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Fang, B. RAS signaling and anti-RAS therapy: Lessons learned from genetically engineered mouse models, human cancer cells, and patient-related studies. Acta Biochim. Biophys. Sin. 2016, 48, 27–38. [Google Scholar] [CrossRef] [PubMed]
- De Roock, W.; Piessevaux, H.; de Schutter, J.; Janssens, M.; de Hertogh, G.; Personeni, N.; Biesmans, B.; van Laethem, J.L.; Peeters, M.; Humblet, Y.; et al. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann. Oncol. 2008, 19, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Scott, N.; Cross, D.; Sagar, P.; Lewis, F.A.; Blair, G.E.; Taylor, G.R.; Dixon, M.F.; Quirke, P. Prognostic value of p53 overexpression and c-Ki-ras gene mutations in colorectal cancer. Gastroenterology 1993, 104, 57–64. [Google Scholar] [CrossRef]
- Vogelstein, B.; Fearon, E.R.; Hamilton, S.R.; Kern, S.E.; Preisinger, A.C.; Leppert, M.; Nakamura, Y.; White, R.; Smits, A.M.; Bos, J.L. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988, 319, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.K.; Law, P.T.; Lee, C.W.; Cho, C.H.; Fan, D.; Wu, K.; Yu, J.; Sung, J.J. MicroRNA in colorectal cancer: From benchtop to bedside. Carcinogenesis 2011, 32, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Saridaki, Z.; Weidhaas, J.B.; Lenz, H.J.; Laurent-Puig, P.; Jacobs, B.; de Schutter, J.; de Roock, W.; Salzman, D.W.; Zhang, W.; Yang, D.; et al. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin. Cancer Res. 2014, 20, 4499–4510. [Google Scholar] [CrossRef] [PubMed]
- Sha, D.; Lee, A.M.; Shi, Q.; Alberts, S.R.; Sargent, D.J.; Sinicrope, F.A.; Diasio, R.B. Association study of the let-7 miRNA-complementary site variant in the 3′ untranslated region of the KRAS gene in stage III colon cancer (NCCTG N0147 Clinical Trial). Clin. Cancer Res. 2014, 20, 3319–3327. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Nakagawa, Y.; Naoe, T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006, 29, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Guo, X.; Zhang, H.; Xiang, Y.; Chen, J.; Yin, Y.; Cai, X.; Wang, K.; Wang, G.; Ba, Y.; et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009, 28, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Kent, O.A.; Mendell, J.T.; Rottapel, R. Transcriptional Regulation of miR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1. Mol. Cancer Res. 2016, 14, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yu, F.; Ma, Y.; Zhao, R.; Chen, X.; Zhu, J.; Zhang, C.Y.; Chen, J.; Zhang, J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1). J. Biol. Chem. 2013, 288, 9508–9518. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M.D.; Boyd, K.L.; Moyo, T.; Mitra, R.; Duszynski, R.; Arrate, M.P.; Chen, X.; Zhao, Z.; Blackwell, T.S.; Andl, T.; et al. MicroRNA-31 initiates lung tumorigenesis and promotes mutant KRAS-driven lung cancer. J. Clin. Investig. 2016, 126, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Oft, M.; Heider, K.H.; Beug, H. TGFβ signaling is necessary for carcinoma cell invasiveness and metastasis. Curr. Biol. 1998, 8, 1243–1252. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Padgett, R.W. Matters of context guide future research in TGFβ superfamily signaling. Sci. Signal. 2015, 8, re10. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.; Arteaga, C.L. Targeting the TGF β signaling network in human neoplasia. Cancer Cell 2003, 3, 531–536. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B.; Wakefield, L.M. The two faces of transforming growth factor β in carcinogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8621–8623. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Fox, J.L.; Hultine, S.; Sundaram, P.; Wang, W.; Liu, Y.Y.; Furth, E.; Enders, G.H.; El-Deiry, W.; Schelter, J.M.; et al. The myc-miR-17~92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent antiangiogenic factors. Cancer Res. 2010, 70, 8233–8246. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Bostrom, A.K.; Impens, F.; Fredlund, E.; van Peer, G.; de Antonellis, P.; von Stedingk, K.; Ghesquiere, B.; Schulte, S.; Dews, M.; et al. The miR-17–92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell 2010, 40, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Li, H.; Wang, L.; Hu, J.; Jin, T.; Wang, J.; Yang, B.B. MicroRNA-17–5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget 2014, 5, 2974–2987. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.; Zou, J.; Dong, Z.; Zeng, Q.; Wu, D.; Liu, L. Up-regulated miR-17 promotes cell proliferation, tumour growth and cell cycle progression by targeting the RND3 tumour suppressor gene in colorectal carcinoma. Biochem. J. 2012, 442, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Paysan, L.; Piquet, L.; Saltel, F.; Moreau, V. RND3 in Cancer: A Review of the Evidence for Tumor Promoter or Suppressor. Mol. Cancer Res. 2016, 14, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Fiorino, A.; Zoni, E.; Crippa, E.; Reid, J.F.; Gariboldi, M.; Pierotti, M.A. The Effects of miR-20a on p21: Two Mechanisms Blocking Growth Arrest in TGF-β-Responsive Colon Carcinoma. J. Cell. Physiol. 2015, 230, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Liu, C.; Yang, J.; Liu, G.; Feng, F.; Tang, J.; Hu, L.; Li, L.; Jiang, F.; Chen, C.; et al. miR-20a triggers metastasis of gallbladder carcinoma. J. Hepatol. 2013, 59, 518–527. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Y.; Lin, S.; Sun, A.; Chen, R.; Ding, Y.; Ding, Y. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 10534–10544. [Google Scholar] [PubMed]
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. miR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015, 13, 252. [Google Scholar] [CrossRef] [PubMed]
- Petrocca, F.; Vecchione, A.; Croce, C.M. Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor β signaling. Cancer Res. 2008, 68, 8191–8194. [Google Scholar] [CrossRef] [PubMed]
- Lasry, A.; Zinger, A.; Ben-Neriah, Y. Inflammatory networks underlying colorectal cancer. Nat. Immunol. 2016, 17, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Terzic, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and colon cancer. Gastroenterology 2010, 138, 2101–2114 e5. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.; Wang, T.C. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell 2009, 15, 79–80. [Google Scholar] [CrossRef] [PubMed]
- Allen, I.C.; Wilson, J.E.; Schneider, M.; Lich, J.D.; Roberts, R.A.; Arthur, J.C.; Woodford, R.M.; Davis, B.K.; Uronis, J.M.; Herfarth, H.H.; et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity 2012, 36, 742–754. [Google Scholar] [CrossRef] [PubMed]
- Roy, I.; Veldkamp, C.T.; Volkman, B.F.; Dwinell, M.B. Chemokines in colitis: MicroRNA control. Gut 2014, 63, 1202–1204. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Wu, Z.; Huang, L.; Qiu, H.; Wang, L.; Li, L.; Yao, L.; Kang, K.; Qu, J.; Wu, Y.; et al. Mulberry fruit prevents LPS-induced NF-κB/pERK/MAPK signals in macrophages and suppresses acute colitis and colorectal tumorigenesis in mice. Sci. Rep. 2015, 5, 17348. [Google Scholar] [CrossRef] [PubMed]
- Saleh, M.; Trinchieri, G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol. 2011, 11, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, F.G.; DuBois, R.N. Connecting COX-2 and Wnt in cancer. Cancer Cell 2006, 9, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Tsujii, M.; Kawano, S.; DuBois, R.N. Cyclooxygenase-2 expression in human colon cancer cells increases metastatic potential. Proc. Natl. Acad. Sci. USA 1997, 94, 3336–3340. [Google Scholar] [CrossRef] [PubMed]
- Peacock, O.; Lee, A.C.; Cameron, F.; Tarbox, R.; Vafadar-Isfahani, N.; Tufarelli, C.; Lund, J.N. Inflammation and miR-21 pathways functionally interact to downregulate PDCD4 in colorectal cancer. PLoS ONE 2014, 9, e110267. [Google Scholar] [CrossRef] [PubMed]
- Allgayer, H. Pdcd4, a colon cancer prognostic that is regulated by a microRNA. Crit. Rev. Oncol. Hematol. 2010, 73, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Yang, Y.; Xia, Y.; Okugawa, Y.; Yang, J.; Liang, Y.; Chen, H.; Zhang, P.; Wang, F.; Han, H.; et al. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut 2016, 65, 1470–1481. [Google Scholar] [CrossRef] [PubMed]
- Oshima, H.; Oshima, M. The inflammatory network in the gastrointestinal tumor microenvironment: Lessons from mouse models. J. Gastroenterol. 2012, 47, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Adachi, O.; Ogawa, T.; Takeda, K.; Akira, S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 1999, 11, 115–122. [Google Scholar] [CrossRef]
- Liu, S.; Sun, X.; Wang, M.; Hou, Y.; Zhan, Y.; Jiang, Y.; Liu, Z.; Cao, X.; Chen, P.; Liu, Z.; et al. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology 2014, 147, 847–859 e11. [Google Scholar] [CrossRef] [PubMed]
- Rokavec, M.; Oner, M.G.; Li, H.; Jackstadt, R.; Jiang, L.; Lodygin, D.; Kaller, M.; Horst, D.; Ziegler, P.K.; Schwitalla, S.; et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014, 124, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Song, L.; Lin, C.; Liu, A.; Lin, X.; Wu, J.; Li, M.; Li, J. Downregulation of miR-138 sustains NF-κB activation and promotes lipid raft formation in esophageal squamous cell carcinoma. Clin. Cancer Res. 2013, 19, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Sen, A.; Most, P.; Peppel, K. Induction of microRNA-138 by pro-inflammatory cytokines causes endothelial cell dysfunction. FEBS Lett. 2014, 588, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.C.; Tsai, L.L.; Wang, M.L.; Yu, C.H.; Lo, W.L.; Chang, Y.C.; Chiou, G.Y.; Chou, M.Y.; Chiou, S.H. miR145 targets the SOX9/ADAM17 axis to inhibit tumor-initiating cells and IL-6-mediated paracrine effects in head and neck cancer. Cancer Res. 2013, 73, 3425–3440. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Liu, D.; Gao, J.; Liu, M.; Liu, S.; Jiang, M.; Liu, Y.; Zheng, D. TRAIL-induced miR-146a expression suppresses CXCR4-mediated human breast cancer migration. FEBS J. 2013, 280, 3340–3353. [Google Scholar] [CrossRef] [PubMed]
- Trifari, S.; Pipkin, M.E.; Bandukwala, H.S.; Aijo, T.; Bassein, J.; Chen, R.; Martinez, G.J.; Rao, A. MicroRNA-directed program of cytotoxic CD8+ T-cell differentiation. Proc. Natl. Acad. Sci. USA 2013, 110, 18608–18613. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Zhou, L.; Mi, Q.S. MicroRNA miR-150 is involved in Vα14 invariant NKT cell development and function. J. Immunol. 2012, 188, 2118–2126. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.E. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle 2009, 8, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Zeki, S.S.; Graham, T.A.; Wright, N.A. Stem cells and their implications for colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Martello, G.; Piccolo, S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 2010, 11, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Ashley, N. Regulation of intestinal cancer stem cells. Cancer Lett. 2013, 338, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Majumdar, A.P. Signaling in colon cancer stem cells. J. Mol. Signal. 2014, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Zhang, J.; Thomson, A.M.; Lim, B.; Rigoutsos, I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008, 455, 1124–1128. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.T.; Xu, Q.; Tong, J.L.; Zhu, M.M.; Nie, F.; Chen, X.; Xiao, S.D.; Ran, Z.H. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br. J. Cancer 2012, 106, 1320–1330. [Google Scholar] [CrossRef] [PubMed]
- Bitarte, N.; Bandres, E.; Boni, V.; Zarate, R.; Rodriguez, J.; Gonzalez-Huarriz, M.; Lopez, I.; Javier Sola, J.; Alonso, M.M.; Fortes, P.; et al. MicroRNA-451 is involved in the self-renewal, tumorigenicity, and chemoresistance of colorectal cancer stem cells. Stem Cells 2011, 29, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Chen, K.Y.; Chen, J.H.; Wang, L.; Walters, J.; Shin, Y.J.; Goerger, J.P.; Sun, J.; Witherspoon, M.; Rakhilin, N.; et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell 2013, 12, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Guilmeau, S. Notch signaling and intestinal cancer. Adv. Exp. Med. Biol. 2012, 727, 272–288. [Google Scholar] [PubMed]
- Fender, A.W.; Nutter, J.M.; Fitzgerald, T.L.; Bertrand, F.E.; Sigounas, G. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J. Cell. Biochem. 2015, 116, 2517–2527. [Google Scholar] [CrossRef] [PubMed]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Goktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011, 17, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Seton-Rogers, S. Epithelial-mesenchymal transition: Untangling EMT’s functions. Nat. Rev. Cancer 2016, 16, 1. [Google Scholar] [CrossRef] [PubMed]
- Gherardi, E.; Birchmeier, W.; Birchmeier, C.; Vande Woude, G. Targeting MET in cancer: Rationale and progress. Nat. Rev. Cancer 2012, 12, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Weinberg, R.A. Epithelial-mesenchymal transition: At the crossroads of development and tumor metastasis. Dev. Cell 2008, 14, 818–829. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Pan, X.; Cobb, G.P.; Anderson, T.A. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007, 302, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Brabletz, T. The ZEB/miR-200 feedback loop—A motor of cellular plasticity in development and cancer? EMBO Rep. 2010, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, H.S.; Li, X.Y.; Lee, I.; Choi, H.S.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Yoon, D.; Fearon, E.R.; et al. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J. Cell Biol. 2011, 195, 417–433. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Hunten, S.; Kaller, M.; Menssen, A.; Gotz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef] [PubMed]
- Jackstadt, R.; Roh, S.; Neumann, J.; Jung, P.; Hoffmann, R.; Horst, D.; Berens, C.; Bornkamm, G.W.; Kirchner, T.; Menssen, A.; et al. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J. Exp. Med. 2013, 210, 1331–1350. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Jackstadt, R.; Siemens, H.; Li, H.; Kirchner, T.; Hermeking, H. p53-induced miR-15a/16–1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2014, 74, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.; Park, S.M.; Radjabi, A.R.; Schickel, R.; Kistner, E.O.; Jewell, D.A.; Feig, C.; Lengyel, E.; Peter, M.E. Let-7 expression defines two differentiation stages of cancer. Proc. Natl. Acad. Sci. USA 2007, 104, 11400–11405. [Google Scholar] [CrossRef] [PubMed]
- King, C.E.; Cuatrecasas, M.; Castells, A.; Sepulveda, A.R.; Lee, J.S.; Rustgi, A.K. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011, 71, 4260–4268. [Google Scholar] [CrossRef] [PubMed]
- Cottonham, C.L.; Kaneko, S.; Xu, L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 2010, 285, 35293–35302. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Y.; Bao, Y.; Yang, W. Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis. Int. J. Mol. Sci. 2017, 18, 890. https://doi.org/10.3390/ijms18040890
Guo Y, Bao Y, Yang W. Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis. International Journal of Molecular Sciences. 2017; 18(4):890. https://doi.org/10.3390/ijms18040890
Chicago/Turabian StyleGuo, Yongchen, Yonghua Bao, and Wancai Yang. 2017. "Regulatory miRNAs in Colorectal Carcinogenesis and Metastasis" International Journal of Molecular Sciences 18, no. 4: 890. https://doi.org/10.3390/ijms18040890