Divergent Expression Patterns and Function Implications of Four nanos Genes in a Hermaphroditic Fish, Epinephelus coioides
Abstract
:1. Introduction
2. Results
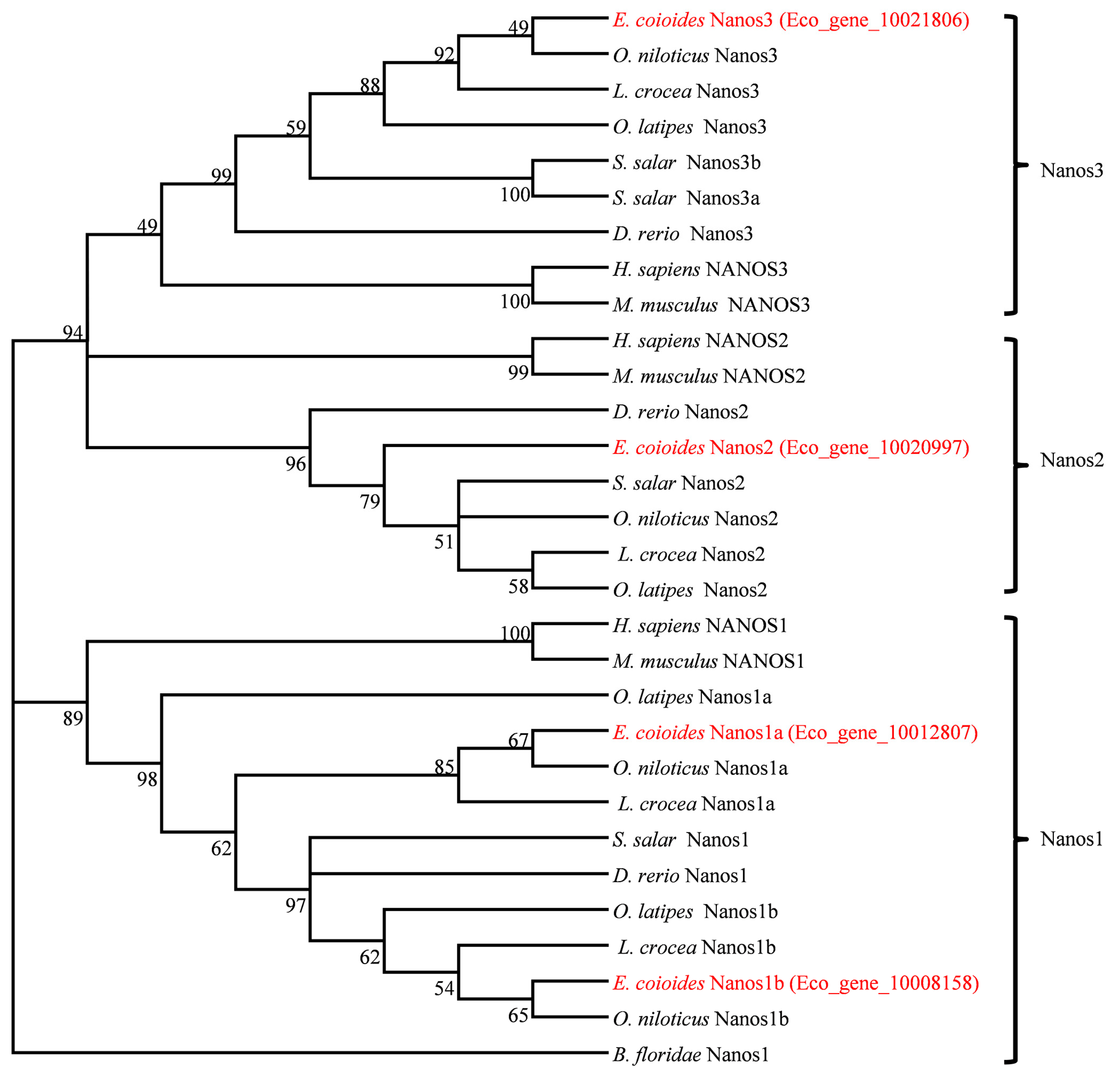
2.1. Molecular Characterization and Phylogenetic Relationship of Four Nanos Genes
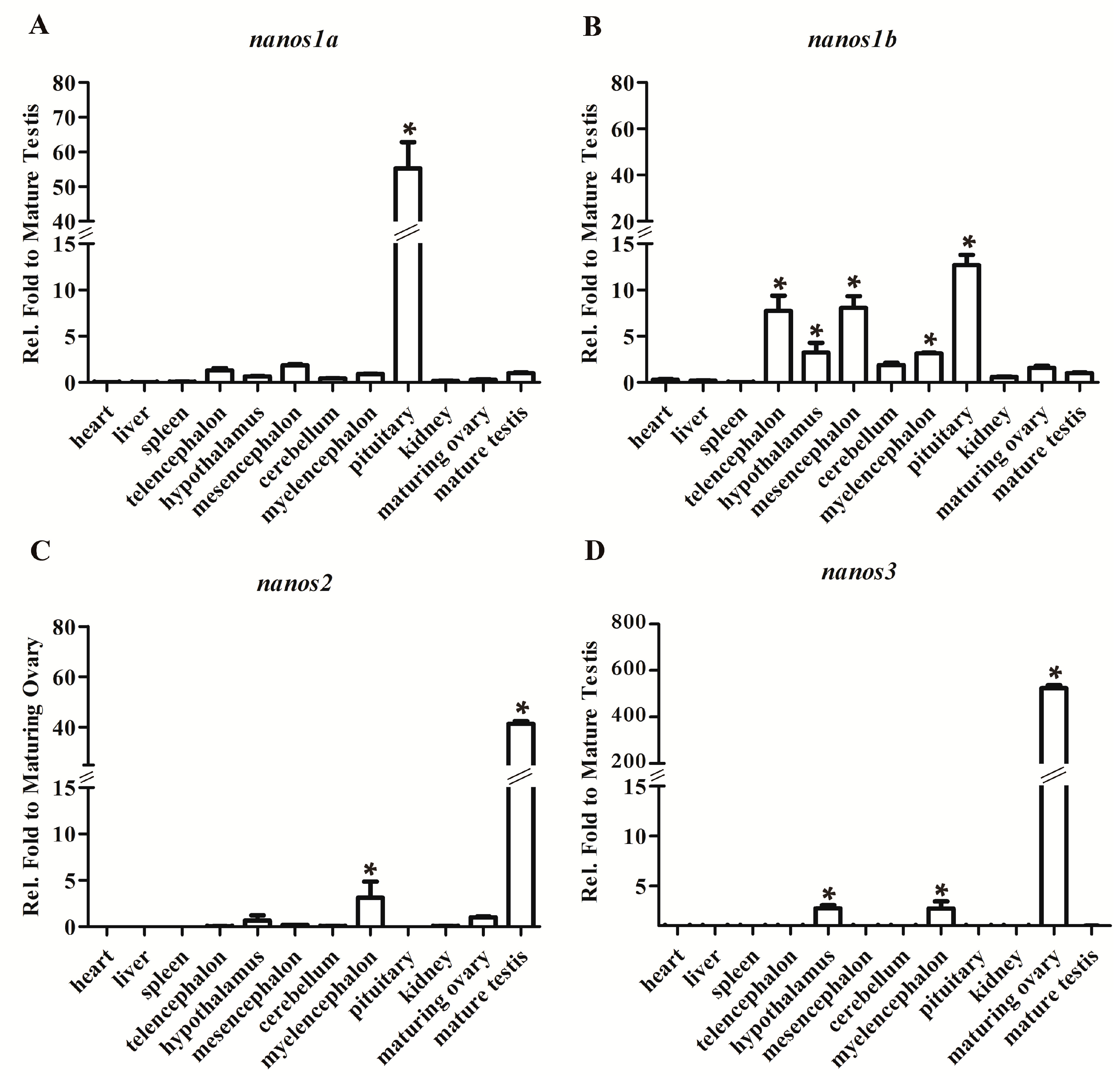
2.2. Differential Expression Patterns of Four Ecnanos Genes in Adult Tissues
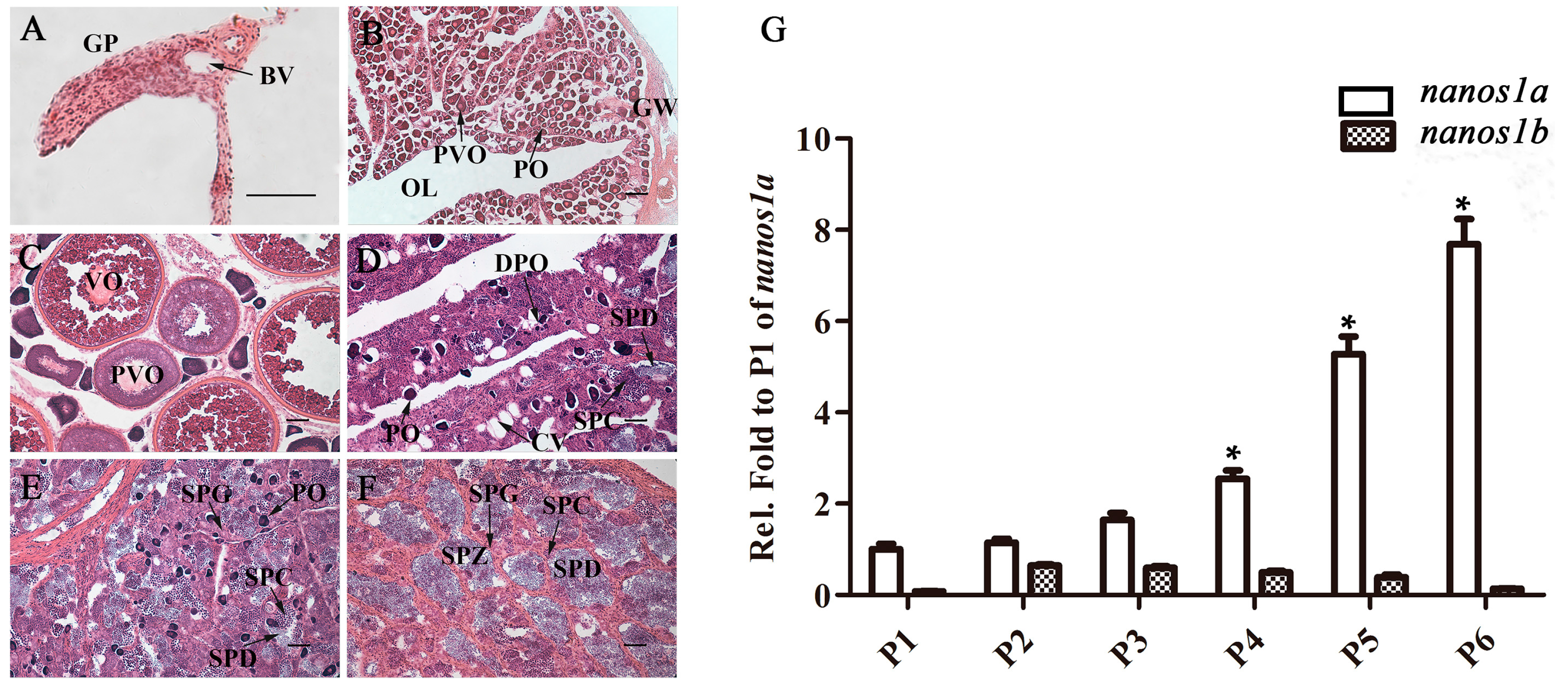
2.3. Ecnanos1a Is Up-Regulated in Pituitaries during Sex Change
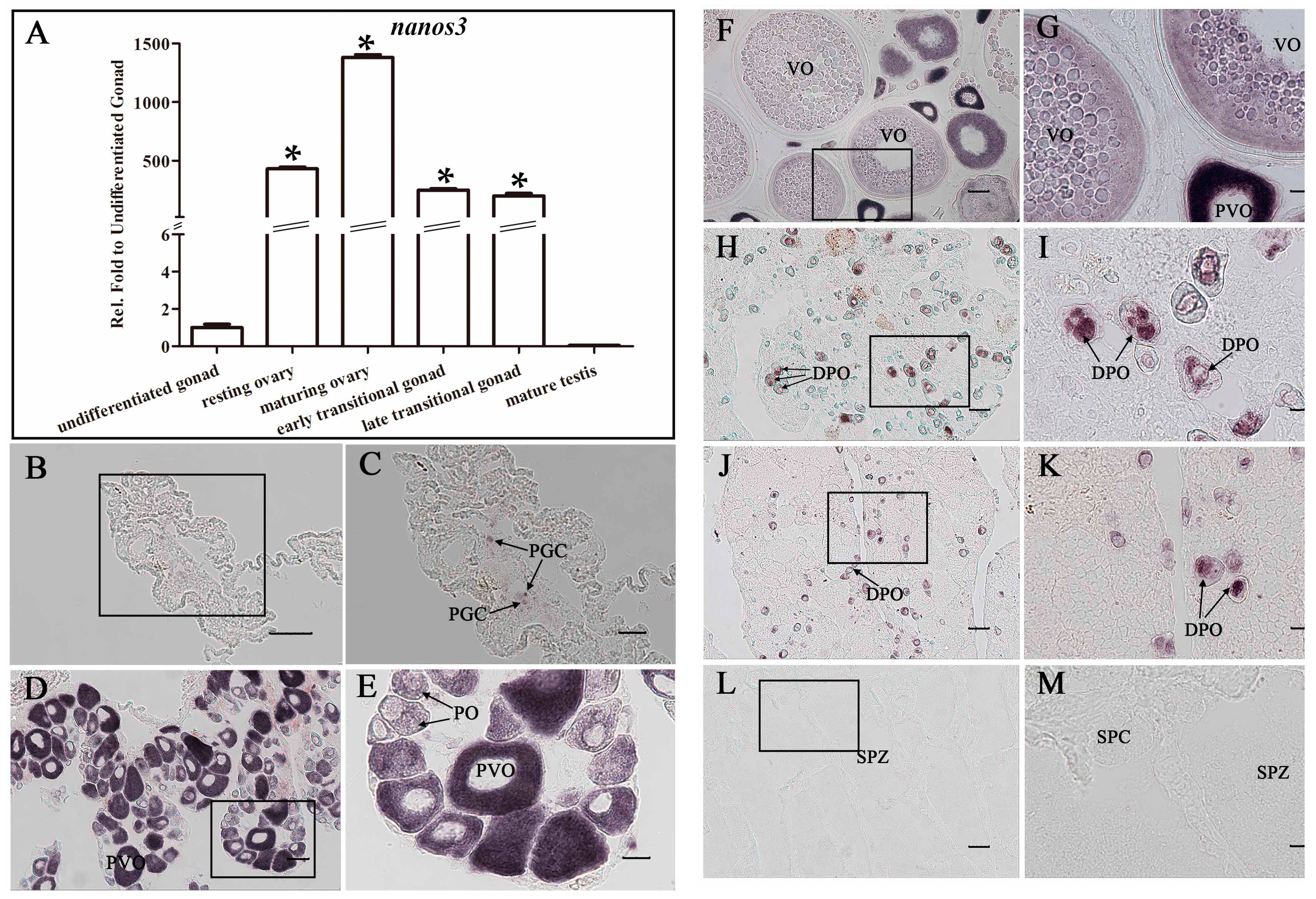
2.4. Ecnanos2 Is Expressed Abundantly in Transitional Gonads and Specifically in Germline Stem Cells
2.5. Ecnanos3 Is Expressed in PGCs and Different Stage Oocytes of Oogenesis
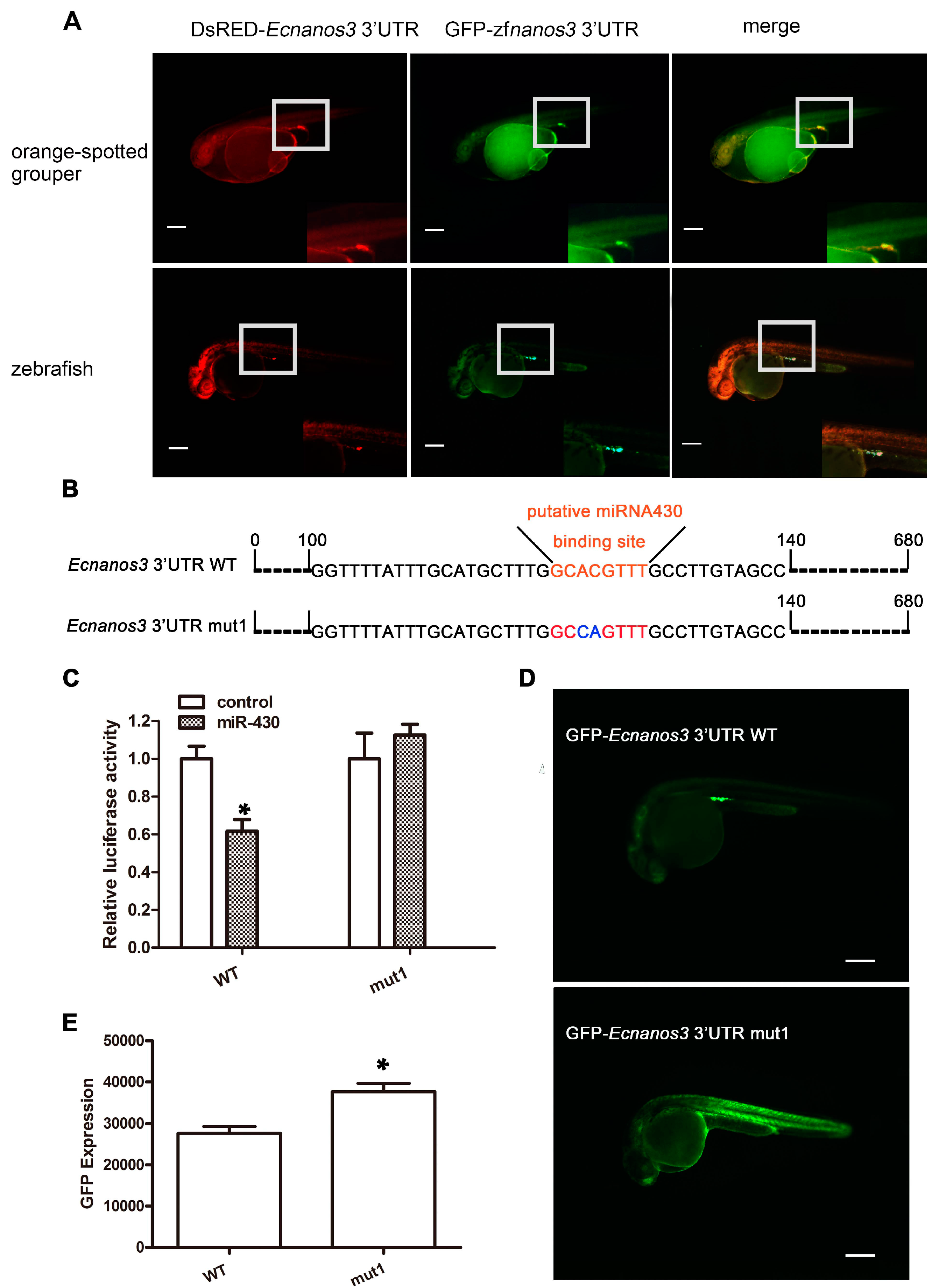
2.6. A Non-Canonical Seed Fragment in Ecnanos3 3′-UTR Is Required for miR-430-Mediated Repression
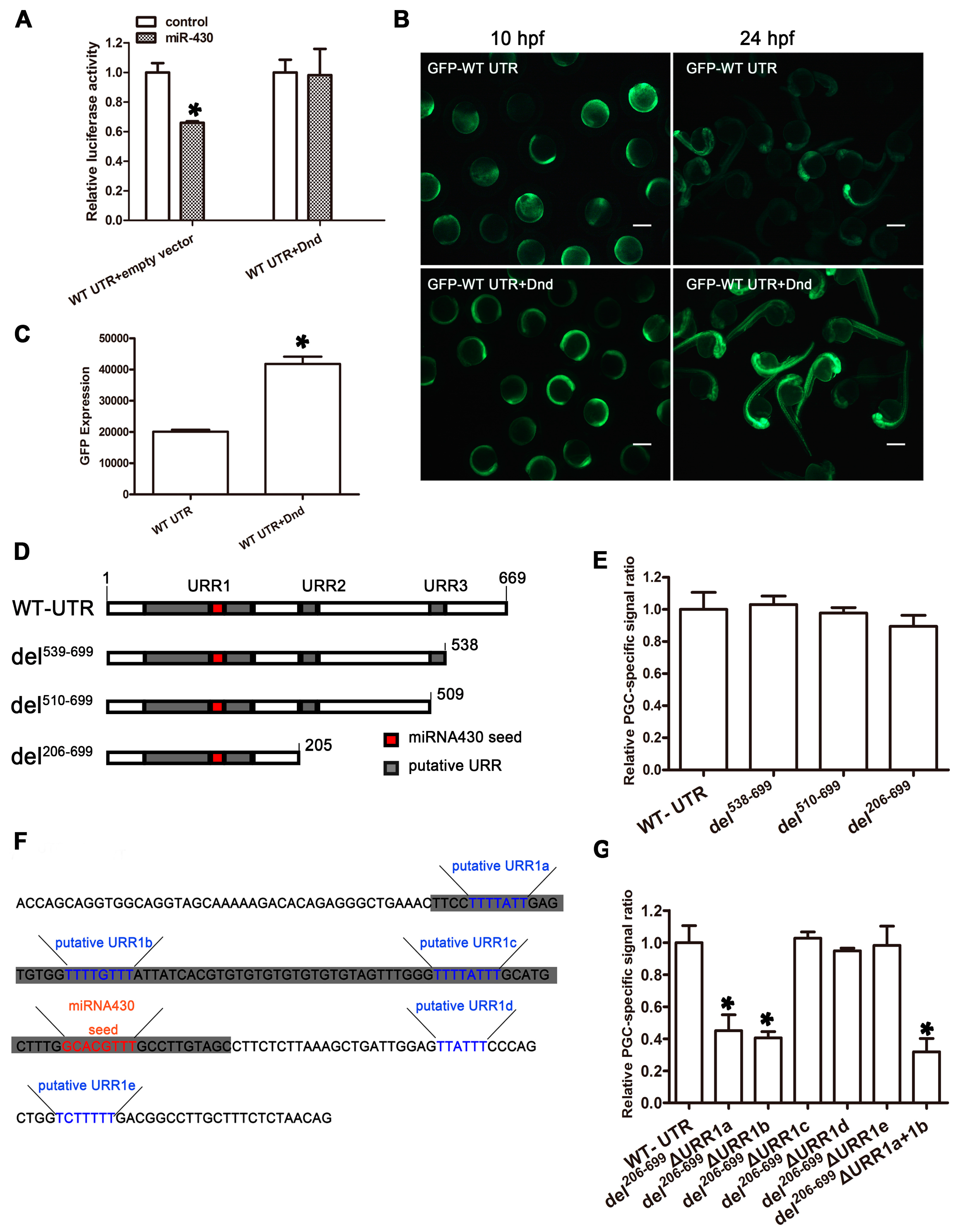
2.7. EcDnd Protects Ecnanos3 from miR-430 Repression in PGCs through a 23 bp U-Rich Element
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
4.3. Sequence and Phylogenetic Analyses
4.4. Histological Examination of Different Gonadal Developmental Stages
4.5. In Situ Hybridization
4.6. RNAs and Microinjection
4.7. Luciferase Reporter Assay
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Xu, H.Y.; Li, M.Y.; Gui, J.F.; Hong, Y.H. Fish germ cells. Sci. China Life Sci. 2010, 53, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, Y.; Yoshizaki, G.; Takeuchi, T. Generation of live fry from intraperitoneally transplanted primordial germ cells in rainbow trout. Biol. Rep. 2003, 69, 1142–1149. [Google Scholar]
- Takeuchi, Y.; Yoshizaki, G.; Takeuchi, T. Biotechnology: Surrogate broodstock produces salmonids. Nature 2004, 430, 629–630. [Google Scholar] [CrossRef] [PubMed]
- Okutsu, T.; Shikina, S.; Kanno, M.; Takeuchi, Y.; Yoshizaki, G. Production of trout offspring from triploid salmon parents. Science 2007, 317, 1517–1517. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Gui, J.; Hong, Y. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev. Dyn. 2005, 233, 872–882. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.X.; Xie, J.L.; Zhou, L.; Hong, Y.H.; Gui, J.F. Evolutionary conservation of dazl genomic organization and its continuous and dynamic distribution throughout germline development in gynogenetic gibel carp. J. Exp. Zool. Part B 2009, 312, 855–871. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, S.Z.; Li, Z.; Wang, Y.; Li, X.Y.; Zhong, J.X.; Zhang, X.J.; Zhang, J.; Zhou, L.; Gui, J.F. Complete depletion of primordial germ cells in an All-female fish leads to sex-biased gene expression alteration and sterile all-male occurrence. BMC Genom. 2015, 16, 971. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lehmann, R. Nanos is the localized posterior determinant in Drosophila. Cell 1991, 66, 637–647. [Google Scholar] [CrossRef]
- Asaoka-Taguchi, M.; Yamada, M.; Nakamura, A.; Hanyu, K.; Kobayashi, S. Maternal pumilio acts together with nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999, 1, 431–437. [Google Scholar] [PubMed]
- Tsuda, M.; Sasaoka, Y.; Kiso, M.; Abe, K.; Haraguchi, S.; Kobayashi, S.; Saga, Y. Conserved role of NANOS proteins in germ cell development. Science 2003, 301, 1239. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Hayashi, M.; Kobayashi, S. Nanos suppresses somatic cell fate in Drosophila germ line. Proc. Natl. Acad. Sci. USA 2004, 101, 10338–10342. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Tsuda, M.; Saga, Y. Functional redundancy among Nanos proteins and a distinct role of nanos2 during male germ cell development. Development 2007, 134, 77–83. [Google Scholar] [PubMed]
- Wang, Z.; Lin, H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science 2004, 303, 2016–2019. [Google Scholar] [CrossRef] [PubMed]
- Julaton, V.T.A.; Pera, R.A.R. Nanos3 function in human germ cell development. Hum. Mol. Genet. 2011, 20, 2238–2250. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, S.; Tsuda, M.; Kitajima, S.; Sasaoka, Y.; Nomura-Kitabayashid, A.; Kurokawa, K.; Saga, Y. Nanos1: A mouse nanos gene expressed in the central nervous system is dispensable for normal development. Mech. Dev. 2003, 120, 721–731. [Google Scholar] [CrossRef]
- Aoki, Y.; Nakamura, S.; Ishikawa, Y.; Tanaka, M. Expression and syntenic analyses of four nanos genes in medaka. Zool. Sci. 2009, 26, 112–118. [Google Scholar] [PubMed]
- Lele Z, K.P.H. The zebrafish as a model system in developmental, toxicological and transgenic research. Biotechnol. Adv. 1996, 14, 57–72. [Google Scholar] [PubMed]
- Grunwald, D.J.; Eisen, J.S. Headwaters of the zebrafish—Emergence of a new model vertebrate. Nat. Rev. Genet. 2002, 3, 711–724. [Google Scholar]
- Beer, R.L.; Draper, B.W. Nanos3 maintains germline stem cells and expression of the conserved germline stem cell gene nanos2 in the zebrafish ovary. Dev. Biol. 2013, 374, 308–318. [Google Scholar] [CrossRef] [PubMed]
- Draper, B.W.; McCallum, C.M.; Moens, C.B. Nanos1 is required to maintain oocyte production in adult zebrafish. Dev. Biol. 2007, 305, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Köprunner, M.; Thisse, C.; Bernard, T.; Erez, R. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001, 15, 2877–2885. [Google Scholar] [PubMed]
- Hashimoto, Y.; Maegawa, S.; Nagai, T.; Yamaha, E.; Suzuki, H.; Yasuda, K.; Inoue, K. Localized maternal factors are required for zebrafish germ cell formation. Dev. Biol. 2004, 268, 152–161. [Google Scholar] [PubMed]
- Raz, E. Primordial germ-cell development: The zebrafish perspective. Nat. Rev. Genet. 2003, 4, 690–700. [Google Scholar] [PubMed]
- Bellaiche, J.; Lareyre, J.J.; Cauty, C.; Yano, A.; Allemand, I.; Le Gac, F. Spermatogonial stem cell quest: Nanos2, marker of a subpopulation of undifferentiated a spermatogonia in trout testis. Biol. Rep. 2014, 90, 79. [Google Scholar]
- Kawakami, Y.; Ishihara, M.; Saito, T.; Fujimoto, T.; Adachi, S.; Arai, K.; Yamaha, E. Cryopreservation of green fluorescent protein (GFP)-labeled primordial germ cells with GFP fused to the 3′-untranslated region of the nanos gene by vitrification of japanese eel (Anguilla japonica) somite stage embryos. J. Anim. Sci. 2012, 90, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Chen, X.; Wei, Q.; Zhou, L.; Liu, T.; Gui, J.; Li, C.; Cao, H. Molecular and expression characterization of a nanos1 homologue in chinese sturgeon, acipenser sinensis. Gene 2012, 511, 285–292. [Google Scholar] [PubMed]
- Li, M.; Tan, X.; Jiao, S.; Wang, Q.; Wu, Z.; You, F.; Zou, Y. A new pattern of primordial germ cell migration in olive flounder (Paralichthys olivaceus) identified using nanos3. Dev. Genes Evol. 2015, 225, 195–206. [Google Scholar] [PubMed]
- Presslauer, C.; Nagasawa, K.; Fernandes, J.M.; Babiak, I. Expression of vasa and nanos3 during primordial germ cell formation and migration in atlantic cod (Gadus morhual). Theriogenology 2012, 78, 1262–1277. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, Y.; Saito, T.; Fujimoto, T.; Goto-Kazeto, R.; Takahashi, E.; Adachi, S.; Arai, K.; Yamaha, E. Visualization and motility of primordial germ cells using green fluorescent protein fused to 3′-UTR of common carp nanos-related gene. Aquaculture 2011, 317, 245–250. [Google Scholar] [CrossRef]
- D’Agostino, I.; Merritt, C.; Chen, P.L.; Seydoux, G.; Subramaniam, K. Translational repression restricts expression of the C. elegans nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 2006, 292, 244–252. [Google Scholar] [PubMed]
- Gavis, E.R.; Lehmann, R. Translation regulation of nanos by RNA localization. Lett. Nat. 1994, 369, 315–318. [Google Scholar]
- Škugor, A.; Slanchev, K.; Torgersen, J.S.; Tveiten, H.; Andersen, O. Conserved mechanisms for germ cell-specific localization of nanos3 transcripts in teleost species with aquaculture significance. Mar. Biotechnol. 2014, 16, 256–264. [Google Scholar] [PubMed]
- Mishima, Y.; Giraldez, A.J.; Takeda, Y.; Fujiwara, T.; Sakamoto, H.; Schier, A.F.; Inoue, K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol. 2006, 16, 2135–2142. [Google Scholar] [CrossRef] [PubMed]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Oude Vrielink, J.A.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Orom, U.A.; et al. RNA-binding protein DND1 inhibits microRNA access to target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [PubMed]
- Giraldez, A.J.; Mishima, Y.; Rihel, J.; Grocock, R.J.; van Dongen, S.; Inoue, K.; Schier, A.F. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 2006, 312, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Slanchev, K.; Stebler, J.; Goudarzi, M.; Cojocaru, V.; Weidinger, G.; Raz, E. Control of dead end localization and activity—Implications for the function of the protein in antagonizing miRNA function. Mech. Dev. 2009, 126, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Spagnoli, F.M.; Brivanlou, A.H. The mir-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 2009, 16, 517–527. [Google Scholar] [PubMed]
- Yeh, S.L.; Dai, Q.C.; Chu, Y.T.; Kuo, C.M.; Ting, Y.Y.; Chang, C.F. Induced sex change, spawning and larviculture of potato grouper, Epinephelus tukula. Aquaculture 2003, 228, 371–381. [Google Scholar]
- Chao, T.M.; Lim, L.C. Recent developments in the breeding of grouper (Epinephelus spp.) in singapore. Singap. J. Prim. Ind. 1991, 19, 78–93. [Google Scholar]
- Yao, B.; Zhou, l.; Wang, Y.; Xia, W.; Gui, J. Differential expression and dynamic changes of SOX3 during gametogenesis and sex reversal in protogynous hermaphroditic fish. J. Exp. Zool. 2007, 307A, 207–219. [Google Scholar]
- Ko, C.F.; Chiou, T.T.; Chen, T.T.; Wu, J.L.; Chen, J.C.; Lu, J.K. Molecular cloning of myostatin gene and characterization of tissue-specific and developmental stage-specific expression of the gene in orange spotted grouper, Epinephelus coioides. Mar. Biotechnol. 2007, 9, 20–32. [Google Scholar] [PubMed]
- Li, W.S.; Chen, D.; Wong, A.O.; Lin, H.R. Molecular cloning, tissue distribution, and ontogeny of mRNA expression of growth hormone in orange-spotted grouper (Epinephelus coioides). Gen. Comp. Endocr. 2005, 144, 78–89. [Google Scholar] [CrossRef]
- Zhou, L.; Gui, J.F. Molecular mechanisms underlying sex change in hermaphroditic groupers. Fish Physiol. Biochem. 2010, 36, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Gui, J.F. Genetic basis and biotechnological manipulation of sexual dimorphism and sex determination in fish. Sci. China Life Sci. 2015, 58, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Zhou, L.; Yao, B.; Li, C.J.; Gui, J.F. Differential and spermatogenic cell-specific expression of dmrt1 during sex reversal in protogynous hermaphroditic groupers. Mol. Cell. Endocr. 2007, 263, 156–172. [Google Scholar]
- Wang, Y.; Zhou, L.; Yao, B.; Li, C.J.; Gui, J.F. Differential expression of thyroid-stimulating hormone β subunit in gonads during sex reversal of orange-spotted and red-spotted groupers. Mol. Cell. Endocr. 2004, 220, 77–88. [Google Scholar]
- Huang, W.; Zhou, L.; Li, Z.; Gui, J.F. Expression pattern, cellular localization and promoter activity analysis of ovarian aromatase (cyp19a1a) in protogynous hermaphrodite red-spotted grouper. Mol. Cell. Endocri. 2009, 307, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.Y.; Zhou, L.; Wang, Y.; Li, Z.; Zhao, J.J.; Zhang, Q.Y.; Gui, J.F. Antibacterial and antiviral roles of a fish β-defensin expressed both in pituitary and testis. PLoS ONE 2010, 5, e12883. [Google Scholar] [CrossRef] [PubMed]
- Marino, G.; Azzurro, E.; Massari, A.; Finoia, M.G.; Mandich, A. Reproduction in the dusky grouper from the southern Mediterranean. J. Fish Biol. 2001, 58, 909–927. [Google Scholar] [CrossRef]
- Weidinger, G.; Stebler, J.; Slanchev, K.; Dumstrei, K.; Wise, C.; Lovell-Badge, R.; Thisse, C.; Thisse, B.; Raz, E. Dead end, a novel vertebrate germ plasm component, is required for zebrafish primordial germ cell migration and survival. Curr. Biol. 2003, 13, 1429–1434. [Google Scholar] [CrossRef]
- Horvay, K.; Claussen, M.; Katzer, M.; Landgrebe, J.; Pieler, T. Xenopus dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev. Biol. 2006, 291, 1–11. [Google Scholar] [PubMed]
- Sakurai, T.; Iguchi, T.; Moriwaki, K.; Noguchi, M. The ter mutation first causes primordial germ cell deficiency in ter/ter mouse embryos at 8 days of gestation. Dev. Growth Differ. 1995, 37, 293–302. [Google Scholar] [CrossRef]
- Aramaki, S.; Sato, F.; Kato, T.; Soh, T.; Kato, Y.; Hattori, M.A. Molecular cloning and expression of dead end homologue in chicken primordial germ cells. Cell Tissue Res. 2007, 330, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tan, X.; Sui, Y.; Jiao, S.; Wu, Z.; You, F. Conserved elements in the nanos3 3′-UTR of olive flounder are responsible for the selective retention of RNA in germ cells. Comp. Biochem. Phys. B 2016, 198, 66–72. [Google Scholar]
- Volff, J.N.; Schartl, M. Evolution of signal transduction by gene and genome duplication in fish. J. Struct. Funct. Genom. 2003, 3, 139–150. [Google Scholar] [CrossRef]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar]
- Pasquier, J.; Cabau, C.; Nguyen, T.; Jouanno, E.; Severac, D.; Braasch, I.; Journot, L.; Pontarotti, P.; Klopp, C.; Postlethwait, J.H.; et al. Gene evolution and gene expression after whole genome duplication in fish: The phylofish database. BMC Genom. 2016, 17, 368. [Google Scholar] [CrossRef] [PubMed]
- Amores, A.; Catchen, J.; Fontenot, Q.; Postlethwait, J.H. Genome evolution and meiotic maps by massively parallel DNA sequencing: Spotted gar, an outgroup for the teleost genome duplication. Genetics 2011, 188, 799–808. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; van de Peer, Y. From 2R to 3R: Evidence for a fish-specific genome duplication (FSGD). BioEssays 2005, 27, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Braasch, I.; Gehrke, A.R.; Smith, J.J.; Kawasaki, K.; Manousaki, T.; Pasquier, J.; Amores, A.; Desvignes, T.; Batzel, P.; Catchen, J. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 2016, 48, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Putnam, N.H.; Butts, T.; Ferrier, D.E.; Furlong, R.F.; Hellsten, U.; Kawashima, T.; Robinson-Rechavi, M.; Shoguchi, E.; Terry, A.; Yu, J.K.; et al. The amphioxus genome and the evolution of the chordate karyotype. Nature 2008, 453, 1064–1071. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, I.; Pfeffer, A.; Friedman, N.; Regev, A. Natural history and evolutionary principles of gene duplication in Fungi. Nature 2007, 449, 54–61. [Google Scholar] [CrossRef]
- Scannell, D.R.; Byrne, K.P.; Gordon, J.L.; Wong, S.; Wolfe, K.H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 2006, 440, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Otto, S.P. The evolutionary consequences of polyploidy. Cell 2007, 131, 452–462. [Google Scholar] [PubMed]
- Lien, S.; Koop, B.F.; Sandve, S.R.; Miller, J.R.; Kent, M.P.; Nome, T.; Hvidsten, T.R.; Leong, J.S.; Minkley, D.R.; Zimin, A.; et al. The atlantic salmon genome provides insights into rediploidization. Nature 2016, 533, 200–205. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, E.D.; Mirarab, S.; Aberer, A.J.; Li, B.; Houde, P.; Li, C.; Ho, S.Y.; Faircloth, B.C.; Nabholz, B.; Howard, J.T. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 2014, 346, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Forbes, A.; Lehmann, R. Nanos and pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 1998, 125, 679–690. [Google Scholar]
- Sada, A.; Suzuki, A.; Suzuki, H.; Saga, Y. The RNA-binding protein nanos2 is required to maintain murine spermatogonial stem cells. Science 2009, 325. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Nagao, I.; Saito, D.; Ebe, Y.; Kinjo, M.; Tanaka, M. Temporal and spatial localization of three germline-specific proteins in medaka. Dev. Dyn. 2008, 237, 800–807. [Google Scholar] [PubMed]
- Saito, T.; Fujimoto, T.; Maegawa, S.; Inoue, K.; Tanaka, M.; Arai, K.; Yamaha, E. Visualization of primordial germ cells in vivo using GFP-nos1 3′-UTR mRNA. Int. J. Dev. Biol. 2006, 50, 691. [Google Scholar] [CrossRef] [PubMed]
- Staton, A.A.; Knaut, H.; Giraldez, A.J. miRNA regulation of sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet. 2011, 43, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Abrams, E.W.; Mullins, M.C. Early zebrafish development: It’s in the maternal genes. Curr.Opin. Genet. Dev. 2009, 19, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Cinalli, R.M.; Rangan, P.; Lehmann, R. Germ cells are forever. Cell 2008, 132, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Oulhen, N.; Yoshida, T.; Yajima, M.; Song, J.L.; Sakuma, T.; Sakamoto, N.; Yamamoto, T.; Wessel, G.M. The 3′-UTR of nanos2 directs enrichment in the germ cell lineage of the sea urchin. Dev. Biol. 2013, 377, 275–283. [Google Scholar] [PubMed]
- Mei, J.; Yue, H.M.; Li, Z.; Chen, B.; Zhong, J.X.; Dan, C.; Zhou, L.; Gui, J.F. C1q-like factor, a target of mir-430, regulates primordial germ cell development in early embryos of carassius auratus. Int. J. Biol. Sci. 2013, 10, 15–24. [Google Scholar]
- Lin, H. The tao of stem cells in the germline. Annu. Rev. Genet. 1997, 31, 455–491. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Kobayashi, K.; Nishimura, T.; Higashijima, S.; Tanaka, M. Identification of germline stem cells in the ovary of the teleost medaka. Science 2010, 328, 1561–1563. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Goto-Kazeto, R.; Arai, K.; Yamaha, E. Xenogenesis in teleost fish through generation of germ-line chimeras by single primordial germ cell transplantation. Biol. Rep. 2008, 78, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, K.; Hasegawa, K.; Chuma, S.; Nakatsuji, N.; Suemori, H. In vitro germ cell differentiation from cynomolgus monkey embryonic stem cells. PLoS ONE 2009, 4, e5338. [Google Scholar]
- Lai, F.; Singh, A.; King, M.L. Xenopus nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development 2012, 139, 1476–1486. [Google Scholar] [CrossRef]
- Lai, F.; Zhou, Y.; Luo, X.; Fox, J.; King, M.L. Nanos1 functions as a translational repressor in the Xenopus germline. Mech. Dev. 2011, 128, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Mosquera, L.; Forristall, C.; Zhou, Y.; Lou King, M. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 1993, 117, 377–386. [Google Scholar] [PubMed]
- Mitrofanov, K.Y.; Zhelankin, A.V.; Shiganova, G.M.; Sazonova, M.A.; Bobryshev, Y.V.; Postnov, A.Y.; Orekhov, A.N. Analysis of mitochondrial DNA heteroplasmic mutations A1555G, C3256T, T3336C, C5178A, G12315A, G13513A, G14459A, G14846A and G15059A in CHD patients with the history of myocardial infarction. Exp. Mol. Pathol. 2016, 100, 87–91. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, M.Q.; Pan, Y.X.; Wu, K.; Xu, Y.H.; Zhang, L.H.; Luo, Z. IRS1 and IRS2: Molecular characterization, tissue expression and transcriptional regulation by insulin in yellow catfish Pelteobagrus fulvidraco. Fish Physiol. Biochem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, gkp335. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic. Acids. Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.H.; Zhou, L.; Wang, Y.; Wang, Z.W.; Li, Z.; Zhang, X.J.; Gui, J.F. Proliferation and resistance difference of a liver-parasitized myxosporean in two different gynogenetic clones of gibel carp. Parasitol. Res. 2014, 113, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Z.H.; Zhou, L.; Li, Z.; Gui, J.F. Grouper tshβ promoter-driven transgenic zebrafish marks proximal kidney tubule development. PLoS ONE 2014, 9, e97806. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, L.; Li, Z.; Li, W.; Gui, J. Apolipoprotein C1 regulates epiboly during gastrulation in zebrafish. Sci. China Life Sci. 2013, 56, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Yue, H.M.; Li, Z.; Wu, N.; Liu, Z.; Wang, Y.; Gui, J.F. Oocyte-specific H2A variant H2AF1O is required for cell synchrony before midblastula transition in early zebrafish embryos. Biol. Rep. 2013, 89, 82. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Liu, M.; Stribinskis, V.; Klinge, C.M.; Ramos, K.S.; Colburn, N.H.; Li, Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 2008, 27, 4373–4379. [Google Scholar] [CrossRef] [PubMed]
- Jing, J.; Xiong, S.; Li, Z.; Wu, J.; Zhou, L.; Gui, J.F.; Mei, J. A feedback regulatory loop involving p53/miR-200 and growth hormone endocrine axis controls embryo size of zebrafish. Sci. Rep. 2015, 5, 15906. [Google Scholar] [CrossRef]
- Sun, F.; Zhang, Y.-B.; Liu, T.-K.; Gan, L.; Yu, F.-F.; Liu, Y.; Gui, J.-F. Characterization of fish IRF3 as an IFN-inducible protein reveals evolving regulation of ifn response in vertebrates. J. Immunol. 2010, 185, 7573–7582. [Google Scholar] [CrossRef]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Z.-H.; Wang, Y.; Lu, W.-J.; Li, Z.; Liu, X.-C.; Li, S.-S.; Zhou, L.; Gui, J.-F. Divergent Expression Patterns and Function Implications of Four nanos Genes in a Hermaphroditic Fish, Epinephelus coioides. Int. J. Mol. Sci. 2017, 18, 685. https://doi.org/10.3390/ijms18040685
Sun Z-H, Wang Y, Lu W-J, Li Z, Liu X-C, Li S-S, Zhou L, Gui J-F. Divergent Expression Patterns and Function Implications of Four nanos Genes in a Hermaphroditic Fish, Epinephelus coioides. International Journal of Molecular Sciences. 2017; 18(4):685. https://doi.org/10.3390/ijms18040685
Chicago/Turabian StyleSun, Zhi-Hui, Yang Wang, Wei-Jia Lu, Zhi Li, Xiao-Chun Liu, Shui-Sheng Li, Li Zhou, and Jian-Fang Gui. 2017. "Divergent Expression Patterns and Function Implications of Four nanos Genes in a Hermaphroditic Fish, Epinephelus coioides" International Journal of Molecular Sciences 18, no. 4: 685. https://doi.org/10.3390/ijms18040685




