Effects of Light-Emitting Diode Irradiation on Growth Characteristics and Regulation of Porphyrin Biosynthesis in Rice Seedlings
Abstract
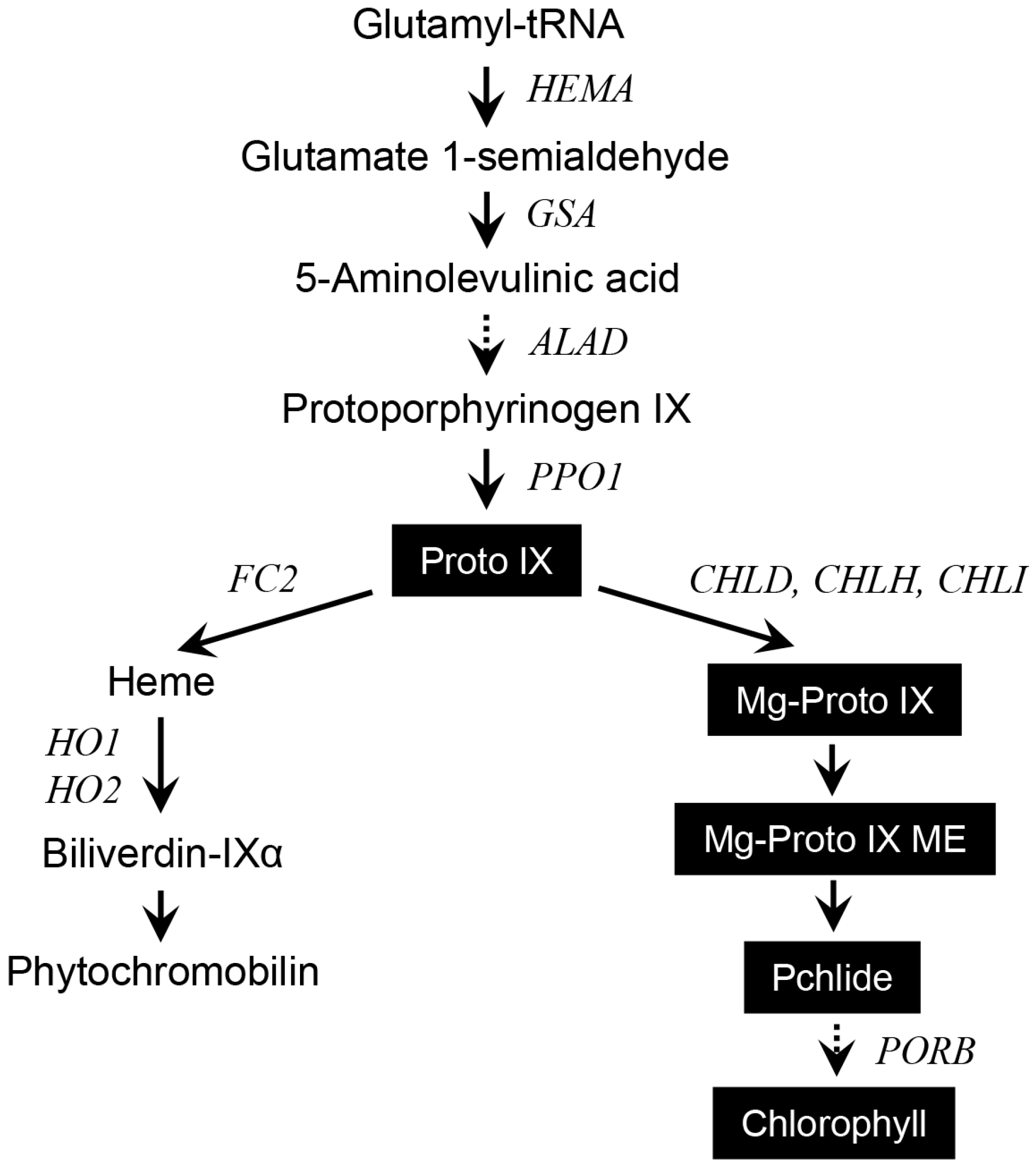
:1. Introduction
2. Results and Discussion
2.1. Effects of Different Wavelengths of LEDs on the Growth and Morphological Appearances of Rice Seedlings
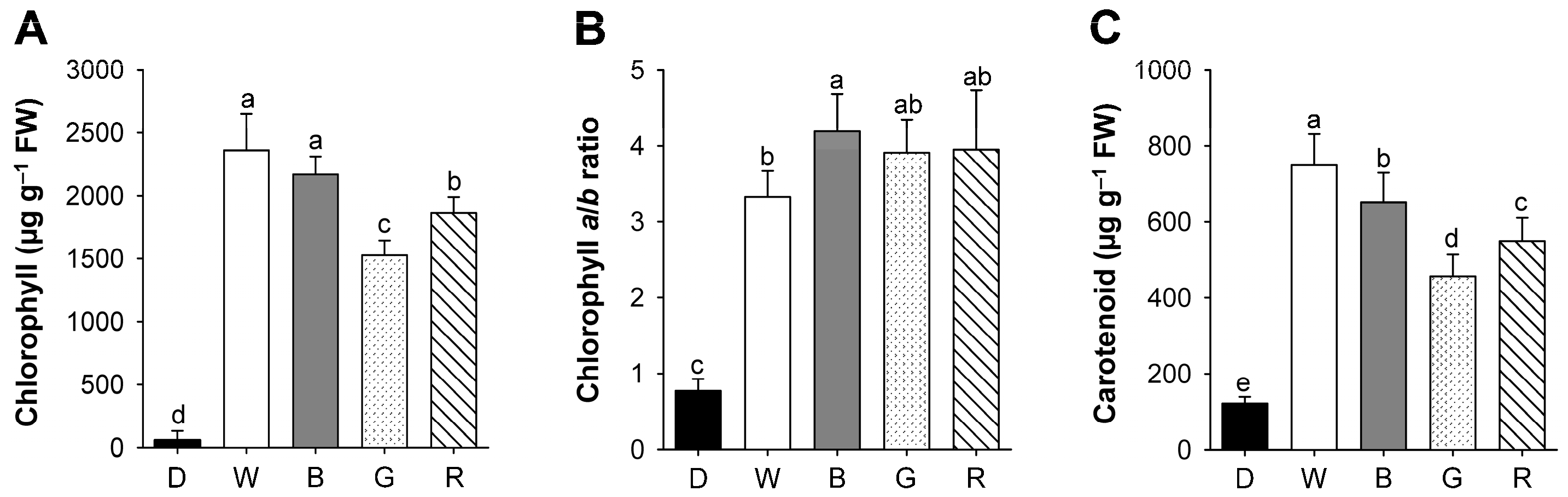
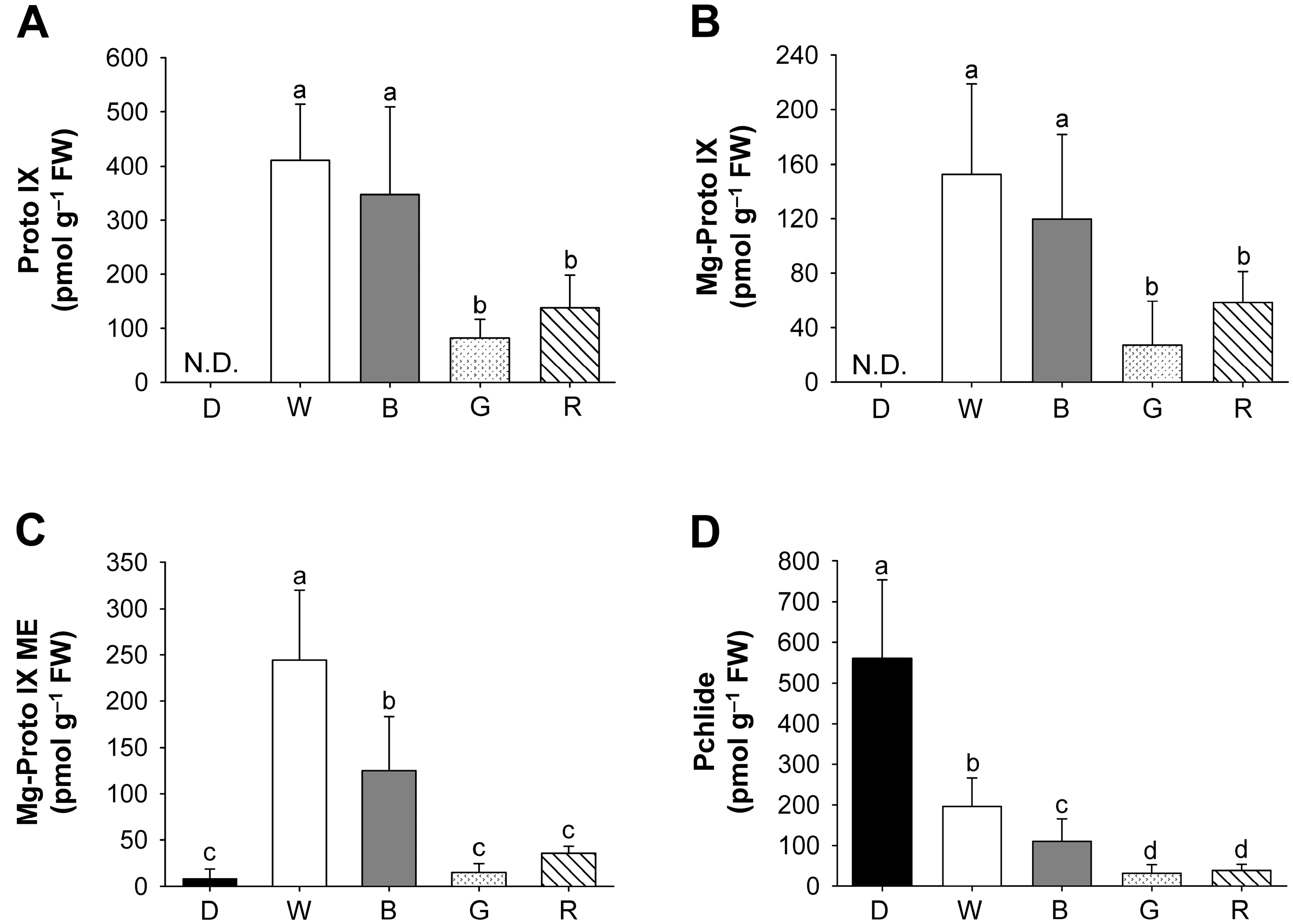
2.2. Effects of Different Wavelengths of LEDs on the Levels of Photosynthetic Pigments and Porphyrin Metabolites
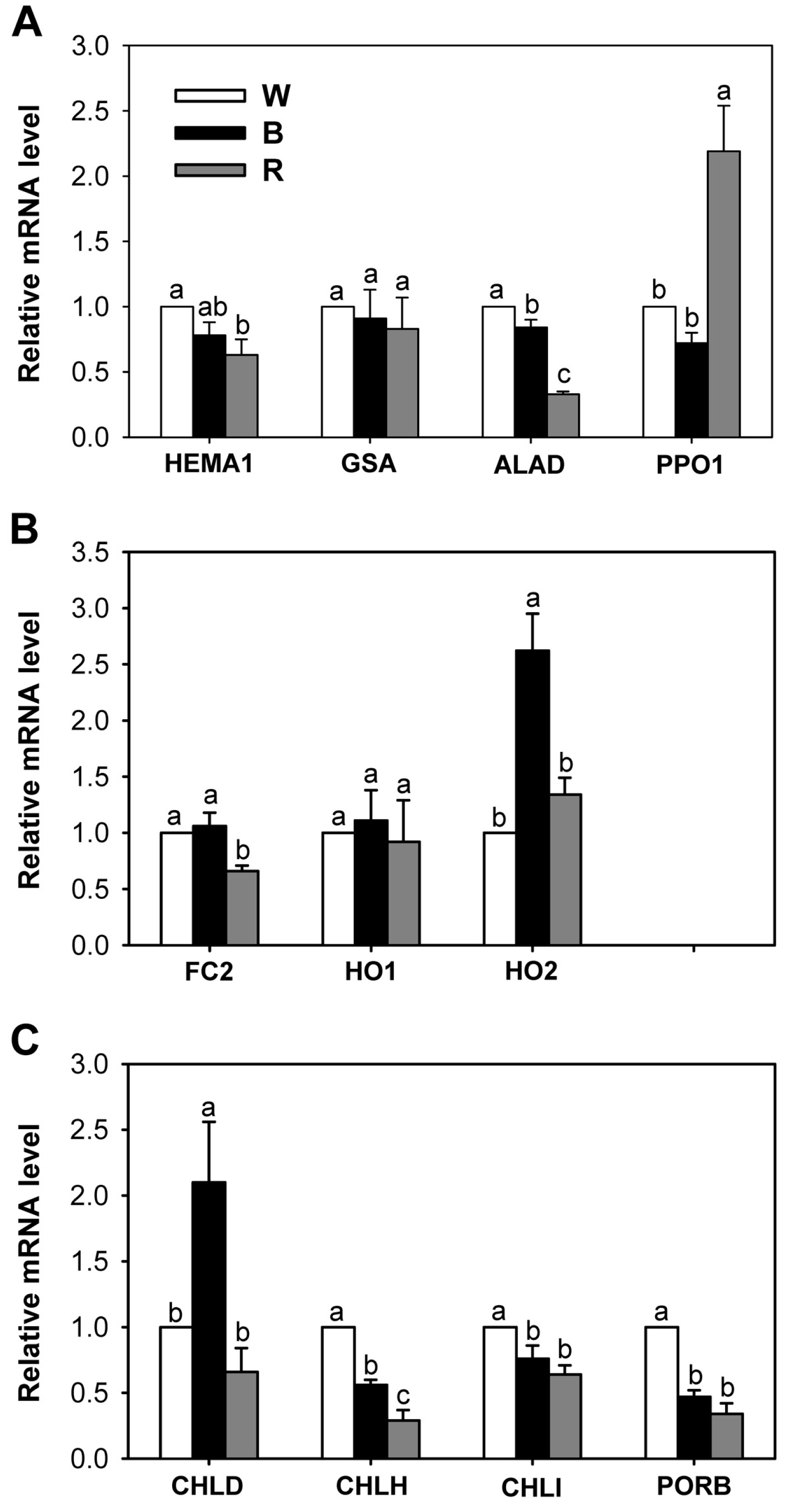
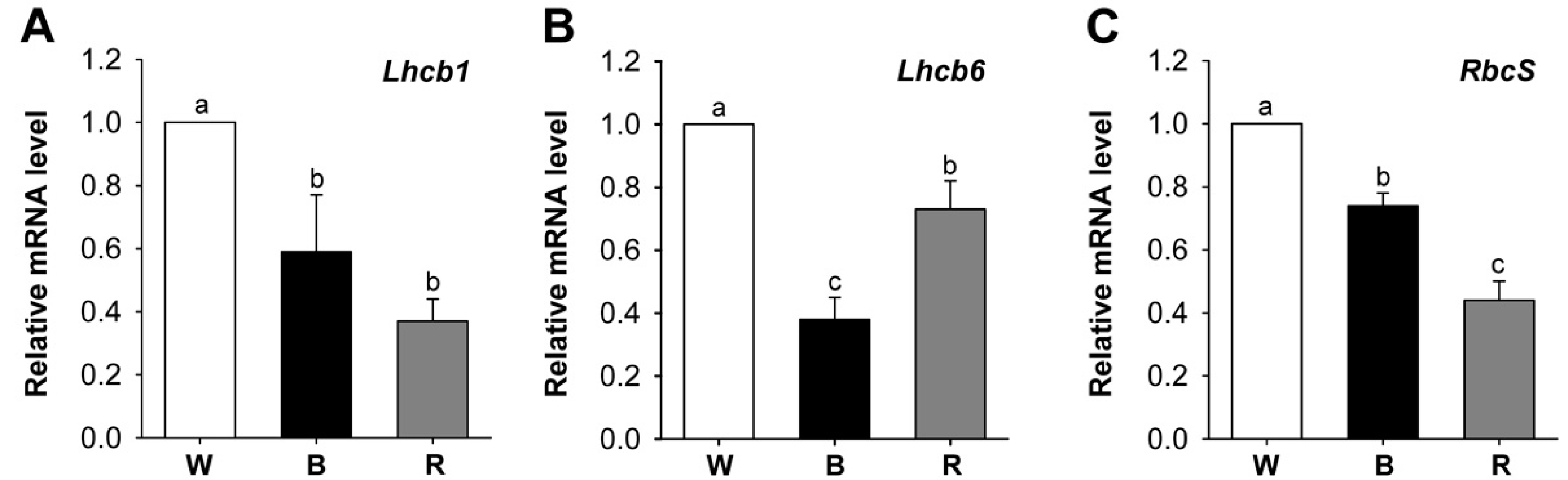
2.3. Different Wavelengths of LEDs Influence Porphyrin Biosynthesis and Retrograde Signaling
3. Materials and Methods
3.1. Plant Growth and Light Treatment Conditions
3.2. Determination of Morphological Characteristics
3.3. Extraction and Analysis of Porphyrin Intermediates and Photosynthetic Pigments
3.4. RNA Extraction and qRT-PCR Analysis
3.5. Data Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Beale, S.I.; Weinstein, J.D. Tetrapyrrole metabolism in photosynthetic organisms. In Biosynthesis of Heme and Chlorophyll; Daily, H.A., Ed.; McGraw-Hill: New York, NY, USA, 1990; pp. 287–391. [Google Scholar]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, S.; Reinbothe, C.; Apel, K.; Lebedev, N. Evolution of chlorophyll biosynthesis—The challenge to survive photooxidation. Cell 1996, 86, 703–705. [Google Scholar] [CrossRef]
- Pham, N.-T.; Kim, J.-G.; Jung, S. Differential antioxidant responses and perturbed porphyrin biosynthesis after exposure to oxyfluorfen and methyl viologen in Oryza sativa. Int. J. Mol. Sci. 2015, 16, 16529–16544. [Google Scholar] [CrossRef] [PubMed]
- Reinbothe, S.; Reinbothe, C. The regulation of enzymes involved in chlorophyll biosynthesis. Eur. J. Biochem. 1996, 237, 323–343. [Google Scholar] [CrossRef] [PubMed]
- Durnford, D.G.; Falkowski, P.G. Chloroplast redox regulation of nuclear gene transcription during photoacclimation. Photosynth Res. 1997, 53, 229–241. [Google Scholar] [CrossRef]
- Murchie, E.H.; Horton, P. Acclimation of photosynthesis to irradiance and spectral quality in British plant species: Chloroplast content, photosynthetic capacity and habitat preference. Plant Cell Environ. 1997, 20, 438–448. [Google Scholar] [CrossRef]
- Plumley, F.G.; Schmidt, G.W. Light-harvesting chlorophyll a/b complexes: Interdependent pigment synthesis and protein assembly. Plant Cell 1995, 7, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.-S.; Chory, J. Signaling between chloroplasts and the nucleus: Can a systems biology approach bring clarity to a complex and highly regulated pathway? Plant Physiol. 2010, 152, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, I.S.; Bondar, V.V.; Bukhov, N.G.; Kotov, A.A.; Kotova, L.M.; Maevskaya, S.N.; Mokronosov, A.T. Effects of light spectral quality on morphogenesis and source-sink relations in radish plants. Russ. J. Plant Physiol. 2001, 48, 415–420. [Google Scholar] [CrossRef]
- Li, Q.; Kubota, C. Effects of supplemental light quality on growth and phytochemicals of baby leaf lettuce. Environ. Exp. Bot. 2009, 67, 59–64. [Google Scholar] [CrossRef]
- Yorio, N.C.; Goins, G.D.; Kagie, H.R.; Wheeler, R.M.; Sager, J.C. Improving spinach, radish, and lettuce growth under red light-emitting diodes (LEDs) with blue light supplementation. Hort. Sci. 2001, 36, 380–383. [Google Scholar]
- Hirose, F.; Inagaki, N.; Hanada, A.; Yamaguchi, S.; Kamiya, Y.; Miyao, A.; Hirochika, H.; Takano, M. Cryptochrome and phytochrome cooperatively but independently reduce active gibberellin content in rice seedlings under light irradiation. Plant Cell Physiol. 2012, 53, 1570–1582. [Google Scholar] [CrossRef] [PubMed]
- Sasakawa, H.; Yamamoto, Y. Effects of blue and red light on unrolling of rice leaves. Planta 1980, 147, 418–421. [Google Scholar] [CrossRef] [PubMed]
- Godo, T.; Fujiwara, K.; Guan, K.; Miyoshi, K. Effects of wavelength of LED-light on in vitro asymbiotic germination and seedling growth of Bletilla ochracea Schltr. (Orchidaceae). Plant Biotechnol. 2011, 28, 397–400. [Google Scholar] [CrossRef]
- Goins, G.D.; Yorio, N.C.; Sanwo, M.M.; Brown, C.S. Photomorphogenesis, photosynthesis, and seed yield of wheat plants under red light-emitting diodes (LEDs) with and without supplemental blue lighting. J. Exp. Bot. 1997, 48, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, K.I.; Doi, M.; Assmann, S.M.; Kinoshita, T. Light regulation of stomatal movement. Annu. Rev. Plant Biol. 2007, 58, 219–247. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-C.; Hou, C.-Y.; Jiang, C.-M.; Wang, Y.-T.; Wang, C.-Y.; Chen, H.-H.; Chang, H.-M. A novel approach of LED light radiation improves the antioxidant activity of pea seedlings. Food Chem. 2007, 101, 1753–1758. [Google Scholar] [CrossRef]
- Senger, H. The effect of blue light on plants and microorganisms. Photochem. Photobiol. 1982, 35, 911–920. [Google Scholar] [CrossRef]
- Lermontova, I.; Grimm, B. Reduced activity of plastid protoporphyrinogen oxidase causes attenuated photodynamic damage during high-light compared to low-light exposure. Plant J. 2006, 48, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, H.-J.; Lee, Y.; Kang, K.; Kim, Y.S.; Grimm, B.; Back, K. Toxic tetrapyrrole accumulation in protoporphyrinogen IX oxidase-overexpressing transgenic rice plants. Plant Mol. Biol. 2008, 67, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Phung, T.-H.; Jung, H.-I.; Park, J.-H.; Kim, J.-G.; Back, K.; Jung, S. Porphyrin biosynthesis control under water stress: Sustained porphyrin status correlates with drought tolerance in transgenic rice. Plant Physiol. 2011, 157, 1746–1764. [Google Scholar] [CrossRef] [PubMed]
- Dai, T.; Gupta, A.; Murray, C.K.; Vrahas, M.S.; Tegos, G.P.; Hamblin, M.R. Blue light for infectious diseases: Propionibacterium acnes, Helicobacter pylori, and beyond? Drug Resist. Updat. 2012, 15, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Casal, J.J. Photoreceptor signaling networks in plant responses to shade. Annu. Rev. Plant Biol. 2013, 64, 403–427. [Google Scholar] [CrossRef] [PubMed]
- Kong, S.G.; Okajima, K. Diverse photoreceptors and light responses in plants. J. Plant Res. 2016, 129, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Muramoto, T.; Tsurui, N.; Terry, M.J.; Yokota, A.; Kohchi, T. Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol. 2002, 130, 1958–1966. [Google Scholar] [CrossRef] [PubMed]
- Terry, M.J.; Linley, P.J.; Kohchi, T. Making light of it: The role of plant heme oxygenases in phytochrome chromophore synthesis. Biochem. Soc. Trans. 2002, 30, 604–609. [Google Scholar] [CrossRef] [PubMed]
- Otterbein, L.E.; Soares, M.P.; Yamashita, K.; Bach, F.H. Heme oxygenase-1: Unleashing the protective properties of heme. Trends Immunol. 2003, 24, 449–455. [Google Scholar] [CrossRef]
- Noriega, G.O.; Balestrasse, K.B.; Batlle, A.; Tomaro, M.L. Heme oxygenase exerts a protective role against oxidative stress in soybean leaves. Biochem. Biophys. Res. Commun. 2004, 323, 1003–1008. [Google Scholar] [CrossRef] [PubMed]
- Yannarelli, G.G.; Noriega, G.O.; Batlle, A.; Tomaro, M.L. Heme oxygenase upregulation in ultraviolet-B irradiated soybean plants involves reactive oxygen species. Planta 2006, 224, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-G.; Back, K.; Lee, H.Y.; Lee, H.-J.; Phung, T.-H.; Grimm, B.; Jung, S. Increased expression of Fe-chelatase leads to increased metabolic flux into heme and confers protection against photodynamically induced oxidative stress. Plant Mol. Biol. 2014, 86, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Gisk, B.; Yasui, Y.; Kohchi, T.; Frankenberg-Dinkel, N. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem. J. 2010, 425, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Strand, A.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 2003, 421, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef] [PubMed]
- Papenbrock, J.; Grimm, B. Regulatory network of tetrapyrrole biosynthesis—Studies of intracellular signaling involved in metabolic and developmental control of plastids. Planta 2001, 213, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Beale, S.I. Enzymes of chlorophyll biosynthesis. Photosyn. Res. 1999, 60, 43–73. [Google Scholar] [CrossRef]
- McCormac, A.C.; Terry, M.J. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 2004, 40, 672–685. [Google Scholar] [CrossRef] [PubMed]
- Lermontova, I.; Grimm, B. Overexpression of plastidic protoporphyrinogen IX oxidase leads to resistance to the diphenyl-ether herbicide acifluorfen. Plant Physiol. 2000, 122, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987, 148, 350–382. [Google Scholar]

| Treatment | Shoot Height (cm) | Leaf Length (cm) | Leaf Width (cm) | Dry Weight (mg·plant−1) |
|---|---|---|---|---|
| 5 days | ||||
| Dark | 17.9 ± 1.6 a | 6.1 ± 1.0 b | 0.19 ± 0.01 d | 11.7 ± 1.3 ab |
| White | 14.3 ± 1.1 c | 5.4 ± 1.0 c | 0.24 ± 0.03 b | 11.6 ± 1.5 abc |
| Blue | 13.6 ± 0.7 d | 7.1 ± 0.9 a | 0.33 ± 0.05 a | 12.0 ± 1.4 a |
| Green | 16.9 ± 1.3 b | 5.8 ± 1.0 ab | 0.22 ± 0.02 c | 10.8 ± 1.9 c |
| Red | 17.7 ± 1.3 a | 6.3 ± 1.2 b | 0.20 ± 0.02 d | 11.1 ± 1.7 bc |
| 10 days | ||||
| Dark | 22.5 ± 3.3 c | 7.4 ± 2.1 d | 0.26 ± 0.03 d | 10.6 ± 1.6 c |
| White | 27.3 ± 3.2 a | 17.0 ± 2.5 a | 0.33 ± 0.03 b | 15.7 ± 2.2 a |
| Blue | 22.4 ± 2.3 c | 13.8 ± 1.5 b | 0.37 ± 0.02 a | 14.9 ± 2.2 a |
| Green | 24.4 ± 1.5 b | 11.8 ± 3.0 c | 0.32 ± 0.03 b | 12.7 ± 2.2 b |
| Red | 27.5 ± 1.7 a | 14.1 ± 2.2 b | 0.28 ± 0.03 c | 12.6 ± 1.3 b |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tran, L.H.; Jung, S. Effects of Light-Emitting Diode Irradiation on Growth Characteristics and Regulation of Porphyrin Biosynthesis in Rice Seedlings. Int. J. Mol. Sci. 2017, 18, 641. https://doi.org/10.3390/ijms18030641
Tran LH, Jung S. Effects of Light-Emitting Diode Irradiation on Growth Characteristics and Regulation of Porphyrin Biosynthesis in Rice Seedlings. International Journal of Molecular Sciences. 2017; 18(3):641. https://doi.org/10.3390/ijms18030641
Chicago/Turabian StyleTran, Lien Hong, and Sunyo Jung. 2017. "Effects of Light-Emitting Diode Irradiation on Growth Characteristics and Regulation of Porphyrin Biosynthesis in Rice Seedlings" International Journal of Molecular Sciences 18, no. 3: 641. https://doi.org/10.3390/ijms18030641





