Localisation Microscopy of Breast Epithelial ErbB-2 Receptors and Gap Junctions: Trafficking after γ-Irradiation, Neuregulin-1β, and Trastuzumab Application
Abstract
:1. Introduction
2. Results
2.1. ErbB Receptor and Connexin-43 Protein Visualization and Measurement by CLSM and LM
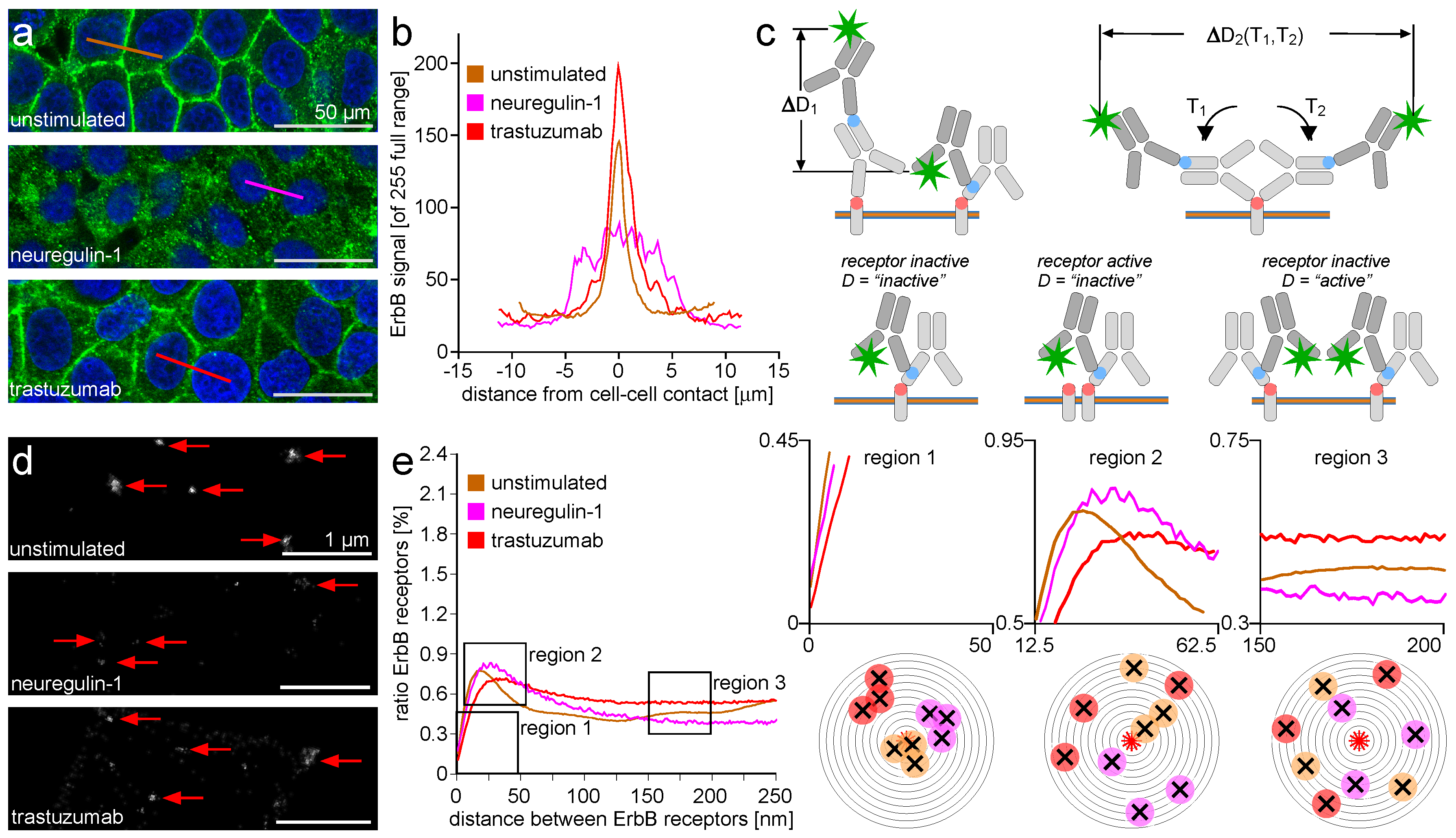
2.2. ErbB Receptor Membrane Clustering Modified by NRG-1 or Trastuzumab as Compared to Unstimulated Situation
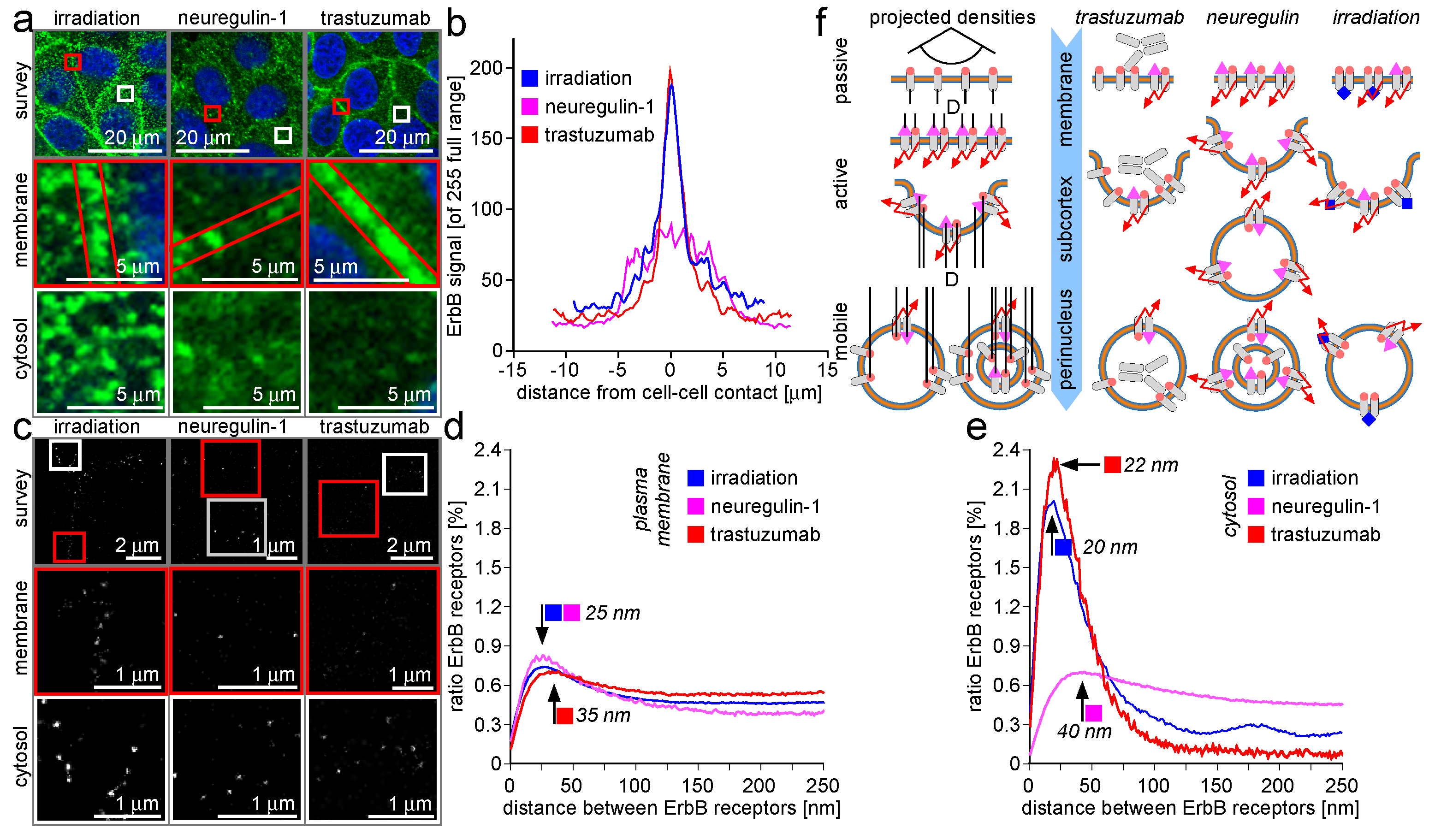
2.3. Trafficking Processes of ErbB Receptors after NRG-1 Stimulation, Trastuzumab Attenuation, and Irradiation
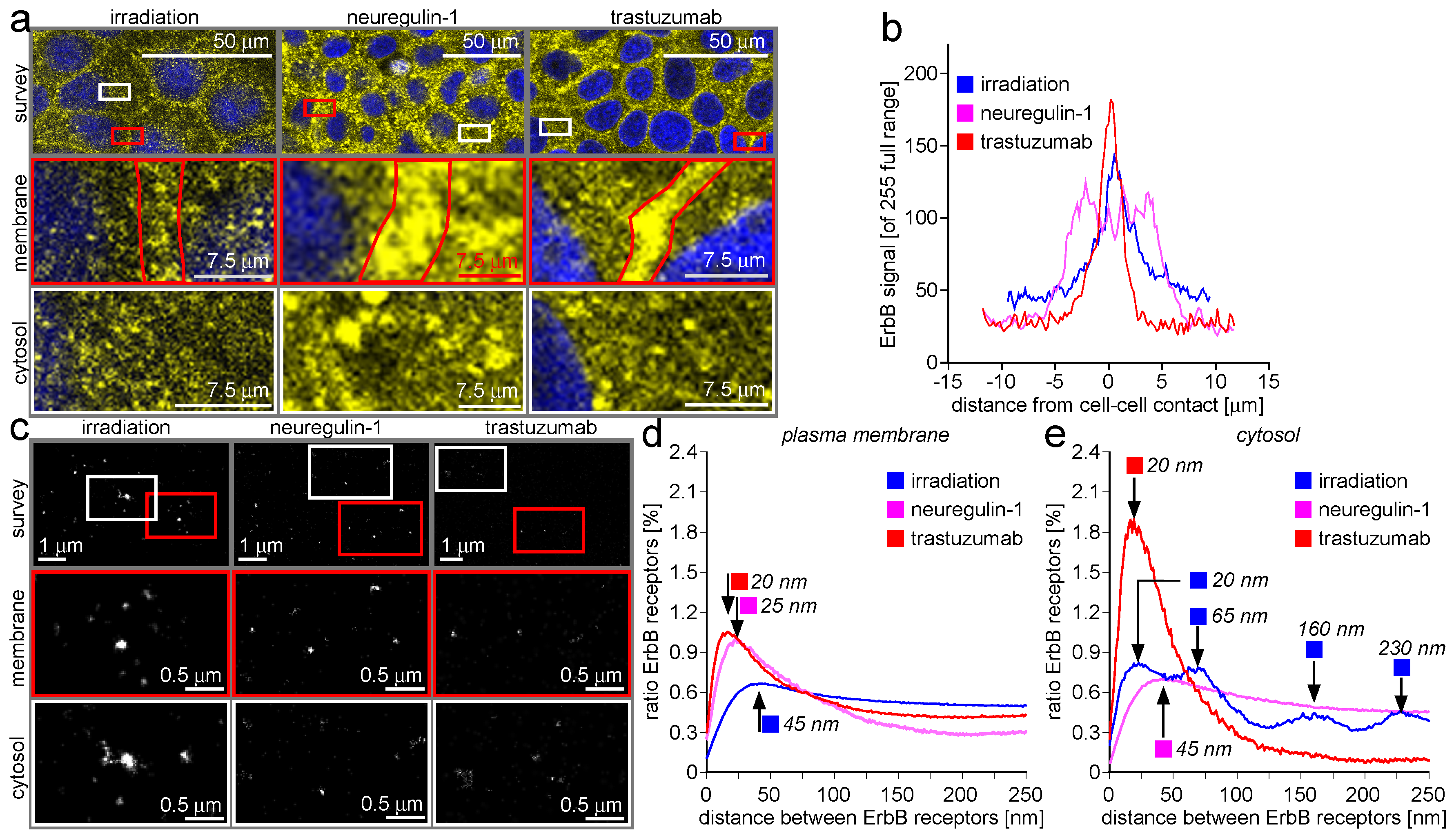
2.4. ErbB-2/3 Receptor Dimers and Gap Junctions Share Common Mobilization Characteristics
3. Discussion
3.1. ErbB Receptor and Connexin-43 Protein Visualization and Measurement by CLSM and LM
3.2. ErbB Receptor Membrane Clustering Modified by NRG-1 or Trastuzumab as Compared to the Unstimulated Situation
3.3. Trafficking Processes of ErbB Receptors after NRG-1 Stimulation, Trastuzumab Attenuation, and Irradiation
3.4. ErbB-2/3 Receptor Dimers and Gap Junctions Share Common Mobilization Characteristics
4. Materials and Methods
4.1. Cell Culture and Cell Manipulation
4.1.1. Cell Culture
4.1.2. Stimulation of MCF-7 Cells with Neuregulin-1
4.1.3. Stimulation of MCF-7 Cells with Trastuzumab
4.1.4. Irradiation of MCF-7 Cells
4.2. Microscopy Specimen Preparation
4.3. Microscopy Data Recording
4.3.1. Confocal Laser Scanning Microscopy (CLSM)
4.3.2. Localization Microscopy (LM)
4.4. Data Processing
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AA(s) | Amino Acid(s) |
| AKT | Gene locus (and protein) family of serine/threonine specific kinases |
| CLSM | Confocal laser scanning microscopy |
| CREB | cAMP response element-binding protein |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DNA-PKs | DNA-dependent protein kinase, catalytic subunit |
| FP(s) | Fluorescent Protein(s) |
| FT | Fourier transform |
| HEPES | N-2-Hydroxyethyl piperazine-N-2-ethane sulphonic acid |
| HSP(s) | Heat Shock Protein(s) |
| IgG | Immuno globulin (of the type) G |
| LM | Localization microscopy |
| MAPK | Mitogen Associated Protein Kinase |
| NPC(s) | Nuclear Pore Complexe(s) |
| NRG-1 | Neuregulin-1 |
| OTF | Optical (or: Objective lens) Transfer Function |
| PALM | Photo Activated Localization Microscopy |
| PKB | Protein Kinase B (Gene product of AKT gene locus) |
| PKCϵ | Protein Kinase C epsilon |
| PI3K | Phosphoinositol Tris Phosphate Kinase |
| ROI(s) | Region(s) of interest |
| ROS | Reactive Oxygen Species |
| RTK(s) | Receptor Tyrosin Kinase(s) |
| STAT | Signal Transducer (and) Activator (of) Transcription |
| STORM | Stochastic Optical Reconstruction Microscopy |
References
- Rhiem, K.; Schmutzler, R.K. Risikofaktoren und prävention des mammakarzinoms. Onkologe 2015, 21, 202–210. [Google Scholar] [CrossRef]
- Szollosi, J.; Balazs, M.; Feuerstein, B.G.; Benz, C.C.; Waldman, F.M. ERBB-2 (HER2-neu) gene copy number, p185HER-2 overexpression, and intratumor heterogeneity in human breast cancer. Cancer Res. 1995, 55, 5400–5407. [Google Scholar] [PubMed]
- Tsuda, H.; Hirohashi, S.; Shimosato, Y.; Hirota, T.; Tsugane, S.; Watanabe, S.; Terada, M.; Yamamoto, H. Correlation between histologic grade of malignancy and copy number of c-erbB-2 gene in breast carcinoma. A retrospective analysis of 176 cases. Cancer 1990, 65, 1794–1800. [Google Scholar] [CrossRef]
- Stern, D.F. ERBB3/HER3 and ERBB2/HER2 duet in mammary development and breast cancer. J. Mammary Gland Biol. Neoplasia 2008, 13, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Saeki, Y.; Endo, T.; Ide, K.; Nagashima, T.; Yumoto, N.; Toyoda, T.; Suzuki, H.; Hayashizaki, Y.; Sakaki, Y.; Okada-Hatakeyama, M. Ligand-specific sequential regulation of transcription factors for differentiation of MCF-7 cells. BMC Genom. 2009, 10, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chun, S.Y.; Kwon, Y.S.; Nam, K.S. Crosstalk between Wnt signaling and Phorbol ester-mediated PKC signaling in MCF-7 human breast cancer cells. Biomed. Pharmacother. 2016, 77, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Brand, T.M.; Iida, M.; Li, C.; Wheeler, D.L. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discov. Med. 2011, 12, 419–432. [Google Scholar] [PubMed]
- Gunzer, K.; Joly, F.; Ferrero, J.M.; Gligorov, J.; Mont-Serrat, H.D.; Uttenreuther-Fischer, M.; Pelling, K.; Wind, S.; Bousquet, G.; Misset, J.L. A phase II study of afatinib, an irreversible ErbB family blocker, added to letrozole in patients with estrogen receptor-positive hormone-refractory metastatic breast cancer progressing on letrozole. Springerplus 2016, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bertelsen, V.; Stang, E. The Mysterious Ways of ErbB2/HER2 Trafficking. Membranes 2014, 4, 424–446. [Google Scholar] [CrossRef] [PubMed]
- Austin, C.D.; Maziere, A.M.D.; Pisacane, P.I.; van Dijk, S.M.; Eigenbrot, C.; Sliwkowski, M.X.; Klumperman, J.; Scheller, R.H. Endocytosis and sorting of ErbB2 and the site of action of cancer therapeutics trastuzumab and geldanamycin. Mol. Biol. Cell 2004, 15, 5268–5282. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.K.; Hung, M.C. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 2015, 282, 3693–3721. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, G.; Liao, H.J. Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 2013, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chou, R.H.; Wang, Y.N.; Hsieh, Y.H.; Li, L. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell 2014, 30, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Hein, A.L.; Ouellette, M.M.; Yan, Y. Radiation-induced signaling pathways that promote cancer cell survival. Int. J. Oncol. 2014, 45, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Britsch, S. The neuregulin-I/ErbB signaling system in development and disease. Adv. Anat. Embryol. Cell Biol. 2007, 190, 1–65. [Google Scholar] [PubMed]
- Montero, J.C.; Seoane, S.; Ocana, A.; Pandiella, A. P-Rex1 participates in Neuregulin-ErbB signal transduction and its expression correlates with patient outcome in breast cancer. Oncogene 2011, 30, 1059–1071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, S.; Zhuang, L.; Zheng, C.Z.; Tengfei, F. Caveolin-1 is involved in radiation-induced ERBB2 nuclear transport in breast cancer cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2012, 32, 888–892. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Yue, J.; Pan, Z.; Wu, H.; Cheng, Y.; Lu, H.; Ren, X.; Yao, M.; Shen, Z.; Yang, J.M. Involvement of Caveolin-1 in repair of DNA damage through both homologous recombination and non-homologous end joining. PLoS ONE 2010, 5, e12055. [Google Scholar] [CrossRef] [PubMed]
- Giri, D.K.; Ali-Seyed, M.; Li, L.Y.; Lee, D.F.; Ling, P.; Bartholomeusz, G.; Wang, S.C.; Hung, M.C. Endosomal transport of ErbB-2: Mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 2005, 25, 11005–11018. [Google Scholar] [CrossRef] [PubMed]
- Cutter, A.R.; Hayes, J.J. A brief review of nucleosome structure. FEBS Lett. 2015, 589, 2914–2922. [Google Scholar] [CrossRef] [PubMed]
- Ozer, G.; Luque, A.; Schlick, T. The chromatin fiber: Multiscale problems and approaches. Curr. Opin. Struct. Biol. 2015, 31, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, Y.; Smerdon, M.J. The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes. J. Biol. Chem. 2013, 288, 13863–13875. [Google Scholar] [CrossRef] [PubMed]
- Avvakumov, N.; Nourani, A.; Cote, J. Histone chaperones: Modulators of chromatin marks. Mol. Cell 2011, 41, 502–514. [Google Scholar] [CrossRef] [PubMed]
- Bowman, G.D.; Poirier, M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef] [PubMed]
- Gurard-Levin, Z.; Zachary, A.; Quivy, J.P.; Almouzni, G. Histone chaperones: Assisting histone traffic and nucleosome dynamics. Annu. Rev. Biochem. 2014, 83, 487–517. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kim, J.H.; Kwon, H. The actin-related protein BAF53 is essential for chromosomal subdomain integrity. Mol. Cells 2015, 38, 789–795. [Google Scholar] [PubMed]
- Oma, Y.; Harata, M. Actin-related proteins localized in the nucleus: From discovery to novel roles in nuclear organization. Nucleus 2011, 2, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Kristo, I.; Bajusz, I.; Bajusz, C.; Borkuti, P.; Vilmos, P. Actin, actin-binding proteins, and actin-related proteins in the nucleus. Histochem. Cell Biol. 2016, 145, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Hu, X.Y.; Xie, X.J.; Xu, Q.Y.; Wang, Y.P.; Liu, X.B.; Xiang, M.X.; Sun, Y.; Wang, J.A. Heat shock protein 90 protects rat mesenchymal stem cells against hypoxia and serum deprivation-induced apoptosis via the PI3K/Akt and ERK1/2 pathways. J. Zheijiang Univ. Sci. B 2010, 11, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Zagouri, F.; Bournakis, E.; Koutsoukos, K.; Papadimitriou, C.A. Heat shock protein 90 (hsp90) expression and breast cancer. Pharmaceuticals 2012, 5, 1008–1020. [Google Scholar] [CrossRef] [PubMed]
- Quanz, M.; Herbette, A.; Sayarath, M.; Koning, L.d.; Dubois, T.; Sun, J.S.; Dutreix, M. Heat shock protein 90α (Hsp90α) is phosphorylated in response to DNA damage and accumulates in repair. J. Biol. Chem. 2012, 287, 8803–8815. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lees-Miller, S.P. Detection and repair of ionizing radiation-induced DNA double strand breaks: New developments in nonhomologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 2013, 86, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.S.; Axelsen, L.N.; Sorgen, P.L.; Verma, V.; Delmar, M.; Holstein-Rathlou, N.H. Gap Junctions. Compr. Physiol. 2012, 2, 1981–2035. [Google Scholar] [PubMed]
- Kessler, E.L.; Boulaksil, M.; van Rijn, H.V.M.; Vos, M.A.; van Veen, T.A.B. Passive ventricular remodeling in cardiac disease: Focus on heterogeneity. Front. Physiol. 2014, 5, 482–496. [Google Scholar] [CrossRef] [PubMed]
- Swietach, P.; Rossini, A.; Spitzer, K.W.; Vaughan-Jones, R.D. H+ ion activation and inactivation of the ventricular gap junction: A basis for spatial regulation of intracellular pH. Circ. Res. 2007, 100, 1045–1054. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Z.; Veenstra, R.D. Monovalent ion selectivity sequences of the rat connexin43 gap junction channel. J. Gen. Physiol. 1997, 109, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Lee, H.H.; Lee, H.J.; Du, Y.Y.; Hirohito, H.; Hung, M.C. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J. Biol. Chem. 2012, 287, 16869–16879. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Ghatnekar, G.S. Cardiac to cancer: Connecting connexins to clinical opportunity. FEBS Lett. 2014, 588, 1349–1364. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Abt, M.A.; Ghatnekar, G.S.; Yeh, E.S. Targeting connexin 43 with α-connexin carboxyl-terminal (ACT1) peptide enhances the activity of the targeted inhibitors, tamoxifen and lapatinib, in breast cancer: Clinical implication for ACT1. BMC Cancer 2015, 15, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Grosely, R.; Sorgen, P.L. A history of gap junction structure: Hexagonal arrays to atomic resolution. Cell Commun. Adhes. 2013, 20, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.M.; Kumar, N.M.; Gilula, N.B. Membrane insertion of gap junction connexins: Polytopic channel forming membrane proteins. J. Cell Biol. 1994, 127, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.M. Biosynthesis and structural composition of gap junction intercellular membrane channels. Eur. J. Cell Biol. 2000, 79, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Lauf, U.; Giepmans, B.N.G.; Lopez, P.; Braconnot, S.; Chen, S.C.; Falk, M.M. Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells. Proc. Natl. Acad. Sci. USA 2002, 99, 10446–10451. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.M.; Fay, A.J.; Puthenveedu, A.M.; Zastrow, M.V.; Jan, Y.N.; Jan, L.Y. Microtubule plus-end-tracking proteins target gap junctions directly from the cell interior to adherens junctions. Cell 2007, 128, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Smyth, J.W.; Vogan, J.M.; Buch, P.J.; Zhang, S.S.; Fong, T.S.; Hong, T.T.; Shaw, R.M. Actin cytoskeleton rest stops regulate anterograde traffic of connexin 43 vesicles to the plasma. Circ. Res. 2012, 110, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Beardslee, M.A.; Laing, J.G.; Beyer, E.C.; Saffitz, J.E. Rapid turnover of connexin43 in the adult rat heart. Circ. Res. 1998, 83, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Kurtenbach, S.; Kurtenbach, S.; Zoidl, G. Gap junction modulation and its implications for heart function. Front. Physiol. 2014, 5, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Dunn, C.A.; Lampe, P.D. Injury-triggered Akt phosphorylation of Cx43: A ZO-1-driven molecular switch that regulates gap junction size. J. Cell Sci. 2014, 127, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Pidoux, G.; Tasken, K. Anchored PKA as a gatekeeper for gap junctions. Commun. Integr. Biol. 2015, 8, e10573611–e10573614. [Google Scholar] [CrossRef] [PubMed]
- Palatinus, J.A.; Rhett, J.M.; Gourdie, R.G. The connexin43 carboxyl terminus and cardiac gap junction organization. Biochim. Biophys. Acta 2012, 1818, 1831–1843. [Google Scholar] [CrossRef] [PubMed]
- Falk, M.M.; Kells, R.M.; Berthoud, V.M. Degradation of connexins and gap junctions. FEBS Lett. 2014, 588, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Kieken, F.; Mutsaers, N.; Dolmatova, E.; Kelly, V.; Wit, A.L.; Kellezi, A.; Hirst-Jensen, B.J.; Duffy, H.S.; Sorgen, P.L. Structural and molecular mechanisms of gap junction remodeling in epicardial border zone myocytes following myocardial infarction. Circ. Res. 2009, 104, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.T.; Nimlamool, W.; Falk, M.M. EGF induces efficient Cx43 gap junction endocytosis in mouse embryonic stem cell colonies via phosphorylation of Ser262, Ser279/282, and Ser368. FEBS Lett. 2014, 588, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.L.; Yusuf, A.M.; Warshaw, D.M. Cargo transport: Molecular motors navigate a complex cytoskeleton. Curr. Opin. Cell Biol. 2008, 20, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Smyth, J.W.; Shaw, R.M. Forward trafficking of ion channels: What the clinician needs to know. Heart Rhythm 2010, 7, 1135–1140. [Google Scholar] [CrossRef] [PubMed]
- Hesketh, G.G.; Shah, M.H.; Halperin, V.L.; Cooke, C.A.; Akar, F.G.; Yen, T.E.; Kass, D.A.; Machamer, C.E.; Eyk, J.E.V.; Tomaselli, G.F. Ultrastructure and regulation of lateralized connexin43 in the failing heart. Circ. Res. 2010, 106, 1153–1163. [Google Scholar] [CrossRef] [PubMed]
- Black, A.T.; Hayden, P.J.; Casillas, R.P.; Heck, D.E.; Gerecke, D.R.; Sinko, P.J.; Laskin, D.L.; Laskin, J.D. Regulation of Hsp27 and Hsp70 expression in human and mouse skin construct models by caveolae following exposure to the model sulfur mustard vesicant, 2-chloroethyl ethyl sulfide. Toxicol. Appl. Pharmacol. 2011, 253, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Mundy, D.I.; Machleidt, T.; Ying, Y.S.; Anderson, R.G.W.; Bloom, G.S. Dual control of caveolar membrane traffic by microtubules and the actin cytoskeleton. J. Cell Sci. 2002, 115, 4327–4339. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Volonte, D.; Galbiati, F. Interaction of caveolin-1 with Ku70 inhibits Bax-mediated apoptosis. PLoS ONE 2012, 7, e39379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dittmann, K.; Mayer, C.; Fehrenbacher, B.; Schaller, M.; Kehlbach, R.; Rodemann, H.P. Nuclear EGFR shuttling induced by ionizing radiation is regulated by phosphorylation at residue Thr654. FEBS Lett. 2010, 584, 3878–3884. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, K.; Mayer, C.; Rodemann, H.P. Nuclear EGFR as novel therapeutic target: Insights into nuclear translocation and function. Strahlenther. Onkol. 2010, 186, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, P.; Pearlman, A.H. EGFR inhibits DNA mismatch repair. Proc. Natl. Acad. Sci. USA 2015, 112, 5556–5557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Hung, M.C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci 2012, 2, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.H.; Zhuo, X.Z.; Ni, Y.J.; Gong, M.; Wang, T.Z.; Lu, Q.; Ma, A.Q. Improvement of cardiac function and reversal of gap junction remodeling by Neuregulin-1β in volume-overloaded rats with heart failure. J. Geriatr. Cardiol. 2012, 9, 172–179. [Google Scholar] [PubMed]
- Wilson, K.L.; Berk, J.M. The nuclear envelope at a glance. J. Cell Sci. 2010, 123, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E. The role of stem cells and gap junctional intercellular communication in carcinogenesis. J. Biochem. Mol. Biol. 2003, 36, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E.; Chang, C.C.; Madhukar, B.V. Modulation of intercellular communication during radiation and chemical carcinogenesis. Radiat. Res. 1990, 123, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Grek, C.L.; Rhett, J.M.; Bruce, J.S.; Ghatnekar, G.S.; Yeh, E.S. Connexin 43, breast cancer tumor suppressor: Missed connections? Cancer Lett. 2016, 374, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C. Recent translational research: Stem cells as the roots of breast cancer. Breast Cancer Res. 2006, 8, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Jiang, W.G.; Mansel, R.E. Gap junctional communication and the tyrosine phosphorylation of connexin 43 in interaction between breast cancer and endothelial cells. Int. J. Mol. Med. 1998, 1, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E. Human adult stem cells as the target cells for the initiation of carcinogenesis and for the generation of “cancer stem cells”. Int. J. Stem Cells 2008, 1, 8–26. [Google Scholar] [CrossRef] [PubMed]
- Sahin, O.; Wang, Q.; Brady, S.W.; Ellis, K.; Wang, H.; Chang, C.C.; Zhang, Q.; Priya, P.; Zhu, R.; Wong, S.T.; et al. Biomarker-guided sequential targeted therapies to overcome therapy resistance in rapidly evolving highly aggressive mammary tumors. Cell Res. 2014, 24, 542–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.H.; Kao, A.P.; Chang, C.C.; Lee, J.N.; Hou, M.F.; Long, C.Y.; Chen, H.S.; Tsai, E.M. Increasing CD44+/CD24− tumor stem cells, and upregulation of COX-2 and HDAC6, as major functions of HER2 in breast tumorigenesis. Mol. Cancer 2010, 9, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Kanczuga-Koda, L.; Sulkowska, M.; Koda, M.; Reszec, J.; Famulski, W.; Baltaziak, M.; Sulkowski, S. Expression of connexin 43 in breast cancer in comparison with mammary dysplasia and the normal mammary gland. Folia Morphol. 2003, 62, 439–442. [Google Scholar]
- Conklin, C.; Huntsman, D.; Yorida, E.; Makretsov, N.; Turbin, D.; Bechberger, J.F.; Sin, W.C.; Naus, C.C. Tissue microarray analysis of connexin expression and its prognostic significance in human breast cancer. Cancer Lett. 2007, 255, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Toulany, M.; Schickfluss, T.A.; Fattah, K.R.; Lee, K.J.; Chen, B.P.C.; Fehrenbacher, B.; Schaller, M.; Chen, D.J.; Rodemann, H.P. Function of erbB receptors and DNA-PKcs on phosphorylation of cytoplasmic and nuclear Akt at S473 induced by erbB1 ligand and ionizing radiation. Radiother. Oncol. 2011, 101, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Baddeley, D.; Cannell, M.B.; Soeller, C. Visualization of localization microscopy data. Microsc. Microanal. 2010, 16, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Godin, A.G.; Lounis, B.; Cognet, L. Super-resolution microscopy approaches for live cell imaging. Biophys. J. 2014, 107, 1777–1784. [Google Scholar] [CrossRef] [PubMed]
- Amgad, M.; Itoh, A.; Tsui, M.M.K. Extending ripleys k-function to quantify aggregation in 2-D grayscale images. PLoS ONE 2015, 10, e0144404. [Google Scholar] [CrossRef] [PubMed]
- Levet, F.; Hosy, E.; Kechkar, A.; Butler, C.; Beghin, A.; Choquet, D.; Sibarita, J.B. SR-Tesseler: A method to segment and quantify localization-based super-resolution microscopy data. Nat. Methods 2015, 12, 1065–1071. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Flores, C.; Nannapaneni, S.; Davidson, K.G.V.; Yasomura, T.; Bennett, M.V.L.; Rash, J.E.; Pereda, A.E. Trafficking of gap junction channels at a vertebrate electrical synapse in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E573–E582. [Google Scholar] [CrossRef] [PubMed]
- Nimlamool, W.; Andrews, R.M.K.; Falk, M.M. Connexin43 phosphorylation by PKC and MAPK signals VEGF-mediated gap junction internalization. Mol. Biol. Cell 2015, 26, 2755–2768. [Google Scholar] [CrossRef] [PubMed]
- Duffy, H.S. The molecular mechanisms of gap junction remodeling. Heart Rhythm 2012, 9, 1331–1334. [Google Scholar] [CrossRef] [PubMed]
- Dittmann, K.; Mayer, C.; Kehlbach, R.; Rodemann, H.P. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport. Mol. Cancer 2008, 7, e1–e9. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, L.; Baldini, G.; Storrie, B. Does super resolution fluorescence microscopy obsolete previous microscopic approaches to protein co-localization. Methods Mol. Biol. 2015, 1270, 255–275. [Google Scholar] [PubMed]
- Tam, J.; Merino, D. Stochastic optical reconstruction microscopy (STORM) in comparison with stimulated emission depletion (STED) and other imaging methods. J. Neurochem. 2015, 135, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Whelan, D.R.; Bell, T.D.M. Image artifacts in single molecule localization microscopy: Why optimization of sample preparation protocols matters. Sci. Rep. 2015, 5, 7924–7934. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, U.; Weiss, M. Hydrophobic mismatch-induced clustering as a primer for protein sorting in the secretory pathway. Biophys. Chem. 2010, 151, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Morozova, D.; Weiss, M.; Guigas, G. Shape as a determinant of membrane protein cluster formation. Soft Matter 2012, 8, 11905–11910. [Google Scholar] [CrossRef]
- Schmidt, U.; Guigas, G.; Weiss, M. Cluster formation of transmembrane proteins due to hydrophobic mismatching. Phys. Rev. Lett. 2008, 101, 128104–128107. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, F.; Arnold, A.M.; Leskovar, K.; Staszek, K.; Fölser, M.; Weghuber, J.; Stockinger, H.; Schütz, G.J. Varying label density allows artifact-free analysis of membrane-protein nanoclusters. Nat. Methods 2016, 13, 661–664. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.M.; Rentero, C.; Rossy, J.; Magenau, A.; Williamson, D.; Rodriguez, M.; Gaus, K. PALM imaging and cluster analysis of protein heterogeneity at the cell surface. J. Biophotonics 2010, 3, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Owen, D.M.; Williamson, D.; Magenau, A.; Gaus, K. Optical techniques for imaging membrane domains in live cells (live-cell palm of protein clustering). Methods Emzymol. 2012, 504, 221–235. [Google Scholar]
- Garcia-Parajo, M.F.; Cambi, A.; Torreno-Pina, J.A.; Thompson, N.; Jacobson, K. Nanoclustering as a dominant feature of plasma membrane organization. J. Cell Sci. 2014, 127, 4995–5005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossy, J.; Pageon, S.V.; Davies, D.M.; Gaus, K. Super-resolution microscopy of the immunological synapse. Curr. Opin. Cell Biol. 2013, 25, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Pageon, S.V.; Tabarin, T.; Yamamoto, Y.; Ma, Y.; Bridgeman, J.S.; Cohnen, A.; Benzing, C.; Gao, Y.; Crowther, M.D.; Tungatt, K.; et al. Functional role of T-cell receptor nanoclusters in signal initiation and antigen discrimination. Proc. Natl. Acad. Sci. USA 2016, 113, e5454–e5463. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.J.; Owen, D.M.; Rossy, J.; Magenau, A.; Wehrmann, M.; Gooding, J.J.; Gaus, K. Pre-existing clusters of the adaptor Lat do not participate in early T cell signaling events. Nat. Immunol. 2011, 12, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Cremer, C.; Kaufmann, R.; Gunkel, M.; Pres, S.; Weiland, Y.; Müller, P.; Ruckelshausen, T.; Lemmer, P.; Geiger, F.; Degenhard, M.; et al. Superresolution imaging of biological nanostructures by spectral precision distance microscopy. Biotechnol. J. 2011, 6, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, M.; Müller, P.; Kaufmann, R.; Cremer, C. Entering the nano-cosmos of the cell by means of spatial position determination microscopy (SPDM): Implications for medical diagnostics and radiation research. IFMBE Proc. 2013, 38, 93–95. [Google Scholar]
- Lemmer, P.; Gunkel, M.; Weiland, Y.; Müller, P.; Baddeley, D.; Kaufmann, R.; Urich, A.; Eipel, H.; Amberger, R.; Hausmann, M.; et al. Using conventional fluorescent markers for far-field fluorescence localization nanoscopy allows resolution in the 10-nm range. J. Microsc. 2009, 235, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Park, E.; Kani, K.; Landgraf, R. Functional isolation of activated and unilaterally phosphorylated heterodimers of ERBB2 and ERBB3 as scaffolds in ligand-dependent signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 13237–13242. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Lu, Y.; Jin, W.; Ang, K.K.; Milas, L.; Fan, Z. Sensitization of breast cancer cells to radiation by trastuzumab. Mol. Cancer Ther. 2003, 2, 1113–1120. [Google Scholar] [PubMed]
- Aertgeerts, K.; Skene, R.; Yano, J.; Sang, B.C.; Zou, H.; Snell, G.; Jennings, A.; Iwamoto, K.; Habuka, N.; Hirokawa, A.; et al. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J. Biol. Chem. 2011, 286, 18756–18765. [Google Scholar] [CrossRef] [PubMed]
- Franklin, M.C.; Carey, K.D.; Vajdos, F.F.; Leahy, D.L.; de Vos, A.M.; Sliwkowski, M.X. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004, 5, 317–328. [Google Scholar] [CrossRef]
- Cho, H.S.; Mason, K.; Ramyar, K.X.; Stanley, A.M.; Gabelli, S.B.; Denney, D.W.D., Jr.; Leahy, D.J. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003, 421, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Ito, A.; Yamamoto, Y.; Sato, M.; Ikeda, K.; Yamamoto, M.; Fujita, H.; Nagamori, E.; Kawabe, Y.; Kamihira, M. Induction of functional tissue-engineered skeletal muscle constructs by defined electrical stimulation. Sci. Rep. 2014, 4, 4781–4788. [Google Scholar] [CrossRef] [PubMed]
- Pereda, A.E.; Curti, S.; Hoge, G.; Cachope, R.; Flores, C.E.; Rash, J.E. Gap junction-mediated electrical transmission: Regulatory mechanisms and plasticity. Biochem. Biophys. Act 2013, 1828, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Endres, N.F.; Engel, K.; Das, R.; Kovacs, E.; Kuriyan, J. Regulation of the catalytic activity of the EGF receptor. Curr. Opin. Struct. Biol. 2011, 21, 777–784. [Google Scholar] [CrossRef] [PubMed]
- James, K.A.; Verkhivker, G.M. Structure-based network analysis of activation mechanisms in the ErbB family of receptor tyrosine kinases: The regulatory spine residues are global mediators of structural stability and allosteric interactions. PLoS ONE 2014, 9, e113488. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.H.; Wang, Y.N.; Hung, M.C. Non-canonical signaling mode of the epidermal growth factor receptor family. Am. J. Cancer Res. 2015, 5, 2944–2958. [Google Scholar] [PubMed]
- Mineev, K.S.; Khabibullina, N.F.; Lyukmanova, E.N.; Dolgikh, D.A.; Kirpichnikov, M.P.; Arseniev, A.S. Spatial structure and dimer-monomer equilibrium of the ErbB3 transmembrane domain in DPC micelles. Biochim. Biophys. Acta 2011, 1808, 2081–2088. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.T.; Tang, F.M.A.; Li, J.J.; Ong, C.; Yung, L.L.Y.; Bay, B.H. Clathrin-Mediated endocytosis of gold nanoparticles in vitro. Anat. Rec. 2015, 298, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Musser, S.M.; Gruenwald, D. Deciphering the structure and function of nuclear pores using single-molecule fluorescence approaches. J. Mol. Biol. 2016, 428, 2091–2119. [Google Scholar] [CrossRef] [PubMed]
- Khan, E.M.; Heidinger, J.M.; Levy, M.; Lisanti, M.P.; Ravid, T.; Goldkorn, T. Epidermal growth factor receptor exposed to oxidative stress undergoes Src- and caveolin-1-dependent perinuclear trafficking. J. Biol. Chem. 2006, 281, 14486–14493. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kuang, J.; Shen, Y.; Majer, M.M.; Nelson, C.C.; Parsawar, K.; Heichman, K.A.; Kuwada, S.K. The atypical histone macroH2A1.2 interacts with HER-2 protein in cancer cells. J. Biol. Chem. 2012, 287, 23171–23183. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, K.; Mahajan, N.P. Cross talk of tyrosine kinases with the DNA damage signaling pathways. Nucleic Acids Res. 2015, 43, 10588–10601. [Google Scholar] [CrossRef] [PubMed]
- Wanner, G.; Mayer, C.; Kehlbach, R.; Rodemann, H.P.; Dittmann, K. Activation of protein kinase C epsilon stimulates DNA-repair via epidermal growth factor receptor nuclear accumulation. Radiother. Oncol. 2008, 86, 383–390. [Google Scholar] [CrossRef] [PubMed]
- Saffitz, J.E.; Davis, L.M.; Darrow, B.J.; Kanter, L.; Laing, J.G.; Beyer, E.C. The molecular basis of anisotropy: Role of gap junctions. J. Cardiovasc. Electrophysiol. 1995, 34, 3576–3588. [Google Scholar] [CrossRef]
- Perkins, G.; Goodenough, D.; Sosinsky, G. Three-dimensional structure of the gap junction connexon. Biophys. J. 1997, 72, 533–544. [Google Scholar] [CrossRef]
- Lin, X.; Gemel, J.; glass, A.; Zemlin, C.W.; Beyer, E.C.; Veenstra, R.D. Connexin40 and connexin43 determine gating properties of atrial gap junction channels. J. Mol. Cell. Cardiol. 2010, 48, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Gakhar, G.; Schrempp, D.; Nguyen, T.A. Regulation of gap junctional intercellular communication by TCDD in HMEC and MCF-7 breast cancer cells. Toxicol. Appl. Pharmacol. 2009, 235, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Leon-Paravic, C.G.; Figueroa, V.A.; Guzman, D.J.; Valderrama, C.F.; Vallejos, A.A.; Fiori, M.C.; Altenberg, G.A.; Reuss, L.; Retamal, M.A. Carbon monoxide (CO) is a novel inhibitor of connexin hemichannels. J. Biol. Chem. 2014, 289, 36150–36157. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.X.; Penuela, S. Connexin and pannexin channels in cancer. BMC Cell Biol. 2015, 17 (Suppl. 1), 105–120. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D. Connexins connection in breast cancer growth and progression. Int. J. Cell Biol. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Teleki, I.; Szasz, A.M.; Maros, M.E.; Gyorffy, B.; Kulka, J.; Meggyeshazi, N.; Kiszner, G.; Balla, P.; Samu, A.; Krenacs, T. Correlations of differentially expressed gap junction connexins Cx26, Cx30, Cx32, Cx43 and Cx46 with breast cancer progression and prognosis. PLoS ONE 2014, 9, e112541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhouk, R.S.; Fares, M.B.; Rahme, G.J.; Hariri, H.H.; Rayess, T.; Dbouk, H.A.; Bazzoun, D.; Al-Labban, D.; El-Sabban, M.E. Context dependent reversion of tumor phenotype by connexin-43 expression in MDA-MB231 cells and MCF-7 cells: Role of β-Catenin/connexin43 association. Exp. Cell Res. 2013, 319, 3065–3080. [Google Scholar] [CrossRef] [PubMed]
- Bazzoun, D.; Lelievre, S.; Talhouk, R. Polarity proteins as regulators of cell junction complexes: Implications for breast cancer. Pharmacol. Ther. 2013, 138, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Kostin, S.; Hein, S.; Bauer, E.P.; Schaper, J. Spatiotemporal development and distribution of intercellular junctions in adult rat cardiomyocytes in culture. Circ. Res. 1999, 85, 154–167. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.G.; Reynhout, J.K.; TenBroek, E.M.; Quade, B.J.; Yasumura, T.; Davidson, K.G.V.; Sheridan, J.D.; Rash, J.E. Gap junction assembly: Roles for the formation plaque and regulation by the C-terminus of connexin43. Mol. Biol. Cell 2012, 23, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Segretain, D.; Falk, M.M. Regulation of connexin biosynthesis, assembly, gap junction formation, and removal. Biochim. Biophys. Acta 2004, 1662, 3–21. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, S.R.; Billaud, M.; Lohman, A.W.; Taddeo, E.P.; Isakson, B.E. Posttranslational modifications in connexins and pannexins. J. Membr. Biol. 2012, 245, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Su, V.; Lau, A.F. Ubiquitination, intracellular trafficking, and degradation of connexins. Arch. Biochem. Biophys. 2012, 524, 16–22. [Google Scholar] [CrossRef]
- Su, V.; Lau, A.F. Connexins: Mechanisms regulating protein levels and intercellular communication. FEBS Lett. 2014, 588, 1212–1220. [Google Scholar] [CrossRef] [PubMed]
- Odiete, O.; Hill, M.F.; Sawyer, D.B. Neuregulin in cardiovascular development and disease. Circ. Res. 2012, 111, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Shaw, R.M. Cardiomyocyte protein trafficking: Relevance to heart disease and opportunities for therapeutic intervention. Trends Cardiovasc. Med. 2015, 25, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.Z.; Xie, Y.; Moyes, K.W.; Gold, J.D.; Askari, B.; Laflamme, M.A. Neuregulin/ErbB signaling regulates cardiac subtype specification in differentiating human embryonic stem cells. Circ. Res. 2010, 107, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Galindo, C.L.; Tran, T.L.; Sawyer, D.B. Neuregulin-1β induces embryonic stem cell cardiomyogenesis via ErbB3/ErbB2 receptors. Biochem. J. 2014, 458, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Barth, A.S.; Zhang, Y.; Li, T.; Smith, R.P.; Chimenti, I.; Terrovitis, I.; Davis, D.R.; Kizana, E.; Ho, A.S.; Rourke, B.O.; et al. Functional impairment of human resident cardiac stem cells by the cardiotoxic antineoplastic agent trastuzumab. Stem Cells Transl. Med. 2012, 1, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Sridharan, V.; Sharma, S.K.; Moros, E.G.; Corry, P.M.; Tripathi, P.; Lieblong, B.J.; Guha, C.; Hauer-Jensen, M.; Boerma, M. Effects of radiation on the epidermal growth factor receptor pathway in the heart. Int. J. Radiat. Biol. 2013, 89, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Manley, S.; Gunzenhäuser, J.; Olivier, N. Cardiotoxicity of concomitant radiotherapy and trastuzumab for early breast cancer. Radiol. Oncol. 2013, 48, 105–112. [Google Scholar]
- Sandoo, A.; Kitas, G.D.; Carmichael, A.R. Breast cancer therapy and cardiovascular risk: Focus on trastuzumab. Vasc. Health Risk Manag. 2015, 11, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Yavas, O.; Yazici, M.; Eren, O.; Oyan, B. The acute effect of trastuzumab infusion on ECG parameters in metastatic breast cancer patients. Swiss Med. Wkly. 2007, 137, 556–558. [Google Scholar]
- Zeglinski, M.; Ludke, A.; Jassal, D.S.; Singal, P.K. Trastuzumab-induced cardiac dysfunction: A dual-hit. Exp. Clin. Cardiol. 2011, 16, 70–74. [Google Scholar] [PubMed]
- Tanriverdi, O.; Meydan, N.; Barutca, S. Long-term effect of trastuzumab on QT dispersion in adjuvant treatment for patients with Her2 receptor positive breast cancer: A pilot study. Med. Oncol. 2012, 29, 3265–3271. [Google Scholar] [CrossRef] [PubMed]
- Rochais, F.; Fischmeister, R. Acute cardiac effects of neuregulin-1/ErbB signaling. Cardiovasc. Res. 2010, 88, 393–394. [Google Scholar] [CrossRef] [PubMed]
- Leithe, E.; Rivedal, E. Epidermal growth factor regulates ubiquitination, internalization and proteasome-dependent degradation of connexin43. J. Cell Sci. 2004, 117, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Galindo, C.L.; Ryzhov, S.; Sawyer, D.B. Neuregulin as a heart failure therapy and mediator of reverse remodeling. Curr. Heart Fail. Rep. 2014, 11, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.S.; Sun, W.; Nomata, K.; Morita, I.; Cruz, A.; Liu, C.J.; Trosko, J.E.; Chang, C.C. Involvement of tyrosine phosphorylation of p185c-erbB2/neu in tumorigenicity induced by X-rays and the neu oncogene in human breast epithelial cells. Mol. Carcinog. 1998, 21, 225–233. [Google Scholar] [CrossRef]
- Hofer, A.; Saez, J.C.; Chang, C.C.; Trosko, J.E.; Spray, D.C.; Dermietzel, R. C-erbB2/neu transfection induces gap junctional communication incompetence in glial cells. J. Neurosci. 1996, 16, 4311–4321. [Google Scholar] [PubMed]
- Trosko, J.E.; Chang, C.C. Mechanism of up-regulated gap junctional intercellular communication during chemoprevention and chemotherapy of cancer. Mutat. Res. 2001, 480–481, 219–229. [Google Scholar] [CrossRef]
- Trosko, J.E.; Chang, C.C. Modulation of cell–cell communication in the cause and chemoprevention/ chemotherapy of cancer. Biofactors 2000, 12, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Pommier, S.J.; Quan, G.G.; Christante, D.; Muller, P.; Newell, A.E.H.; Olson, S.B.; Diggs, B.; Muldoon, L.; Neuwelt, E.; Pommier, R.F. Characterizing the HER2/neu status and metastatic potential of breast cancer stem/progenitor cells. Ann. Surg. Oncol. 2010, 17, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Rakib, M.A.; Lee, W.S.; Kim, G.S.; Han, J.H.; Kim, J.O.; Ha, Y.L. Antiproliferative action of conjugated linoleic acid on human MCF-7 breast cancer cells mediated by enhancement of gap junctional intercellular communication through inactivation of NF-κ B. Evid.-Based Complement. Altern. Med. 2013, 2013. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.S.; Wilson, M.R.; Hayashi, T.; Chang, C.C.; Trosko, J.E. Inhibition of gap junctional intercellular communication in normal human breast epithelial cells after treatment with pesticides, PCBs, and PBBs, alone or in mixtures. Environ. Health Perspect. 1996, 104, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.H.; Kao, A.P.; Lin, T.C.; Chang, C.C.; Kuo, T.C. Promotion of epithelial-mesenchymal transition and tumor growth by 17β-estradiol in an ER+/HER2+ cell line derived from human breast epithelial stem cells. Biotechnol. Appl. Biochem. 2012, 59, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Trosko, J.E.; Chang, C.C. Role of stem cells and gap junctional intercellular communication in human carcinogenesis. Radiat. Res. 2001, 1, 175–180. [Google Scholar] [CrossRef]
- Trosko, J.E.; Chang, C.C.; Madhukar, B.V. Cell-to-cell communication: Relationship of stem cells to the carcinogenic process. Progr. Clin. Biol. Res. 1990, 331, 259–276. [Google Scholar]
- Matesic, D.F.; Ali, A.; Sidorova, T.S.; Burns, T.J. A cell–cell communication marker for identifying targeted tumor therapies. Curr. Bioact. Compd. 2013, 9, 255–262. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilarczyk, G.; Nesnidal, I.; Gunkel, M.; Bach, M.; Bestvater, F.; Hausmann, M. Localisation Microscopy of Breast Epithelial ErbB-2 Receptors and Gap Junctions: Trafficking after γ-Irradiation, Neuregulin-1β, and Trastuzumab Application. Int. J. Mol. Sci. 2017, 18, 362. https://doi.org/10.3390/ijms18020362
Pilarczyk G, Nesnidal I, Gunkel M, Bach M, Bestvater F, Hausmann M. Localisation Microscopy of Breast Epithelial ErbB-2 Receptors and Gap Junctions: Trafficking after γ-Irradiation, Neuregulin-1β, and Trastuzumab Application. International Journal of Molecular Sciences. 2017; 18(2):362. https://doi.org/10.3390/ijms18020362
Chicago/Turabian StylePilarczyk, Götz, Ines Nesnidal, Manuel Gunkel, Margund Bach, Felix Bestvater, and Michael Hausmann. 2017. "Localisation Microscopy of Breast Epithelial ErbB-2 Receptors and Gap Junctions: Trafficking after γ-Irradiation, Neuregulin-1β, and Trastuzumab Application" International Journal of Molecular Sciences 18, no. 2: 362. https://doi.org/10.3390/ijms18020362







