Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS)
Abstract
:1. Introduction
2. Results and Discussion
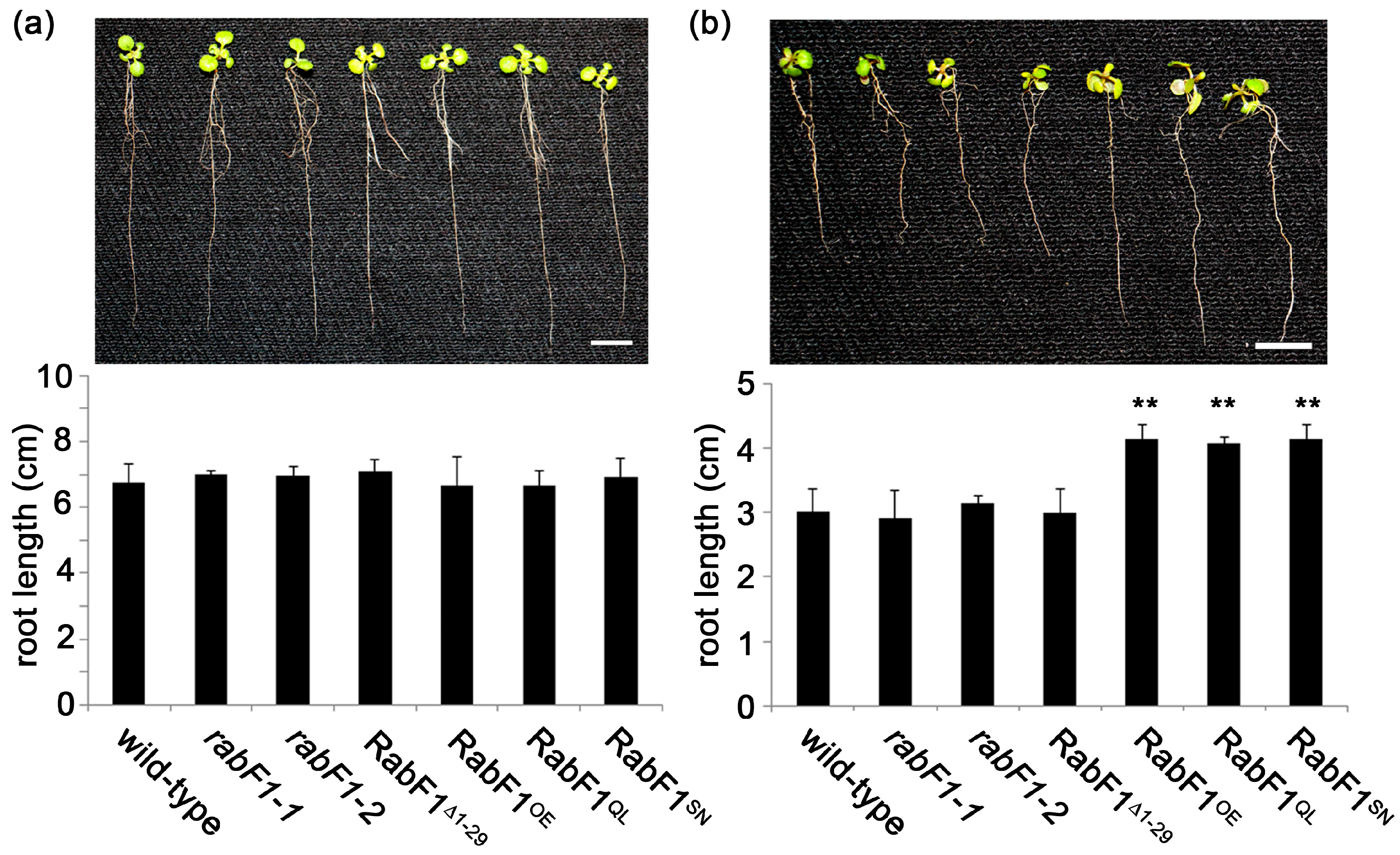
2.1. RabF1OE, RabF1Q93L and RabF1S47N Lines Are More Tolerant to Salt Stress Compared to Wild-Type, rabF1 and RabF1Δ1−29
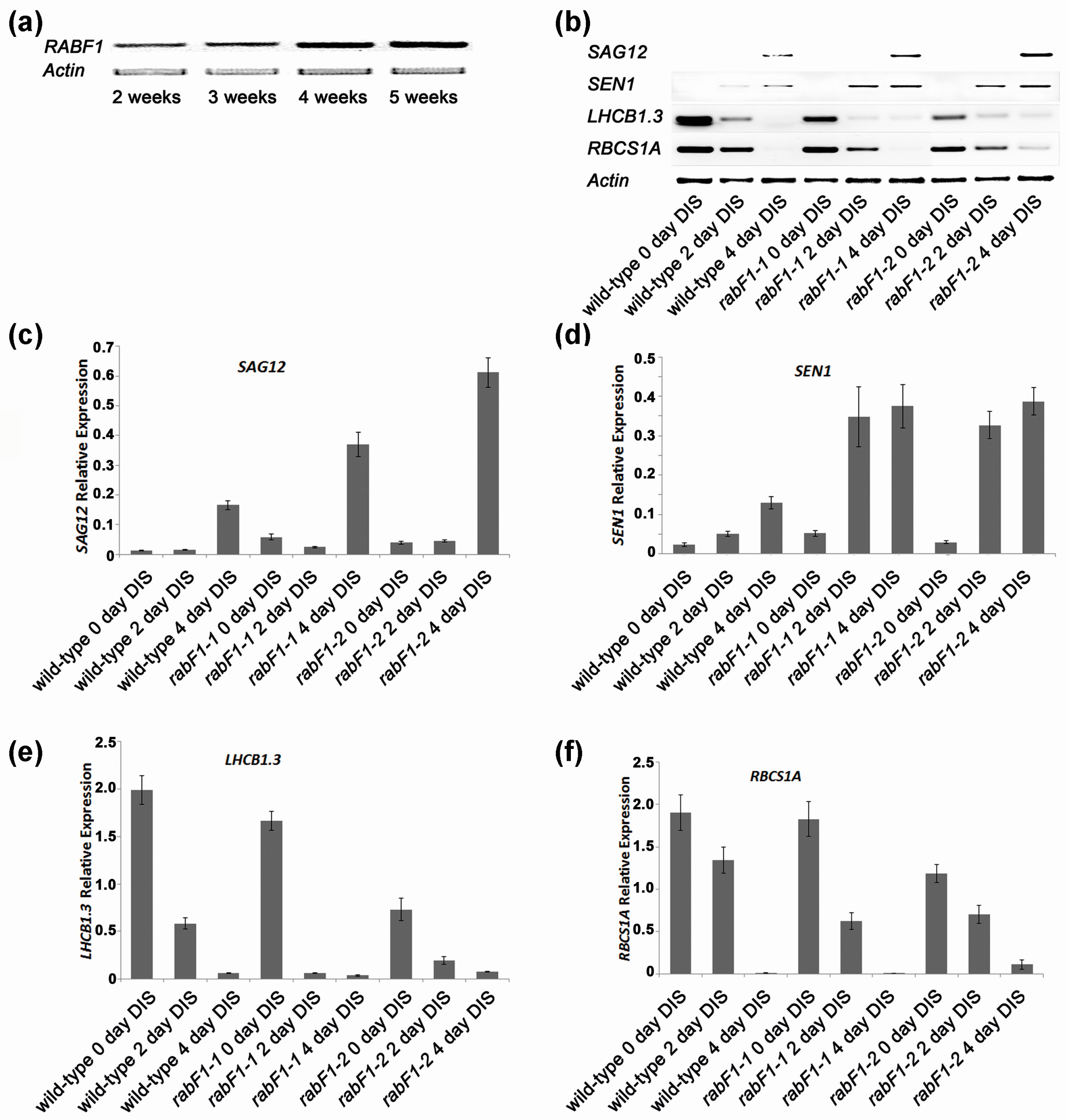
2.2. RabF1 Is Involved in Leaf Senescence
2.3. RabF1 Plays an Important Role in Dark-Induced Senescence (DIS)
2.4. Conclusions
3. Materials and Methods
3.1. Plant Material
3.2. Total RNA Isolation, RT-PCR and Semi-Quantitative RT-PCR
3.3. RabF1 Cloning
3.4. Expression of 35S::RabF1-EYFP (RabF1OE), 35S::RabF1Q93L-EYFP (RabF1Q93L), 35S::RabF1S47N-EYFP (RabF1S47N), 35S::RabF1Δ1−29-EYFP (RabF1Δ1−29) in rabF1
3.5. Plant Growth for Salt Stress
3.6. Dark-Induced Senescence (DIS), Chlorophyll and Conductivity Measurements
Supplementary Materials
Acknowledgment
Author Contributions
Conflicts of Interest
References
- Pfeffer, S.; Aivazian, D. Targeting RAB GTPases to distinct membrane compartments. Nat. Rev. Mol. Cell Biol. 2004, 5, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol. 2001, 11, 487–491. [Google Scholar] [CrossRef]
- Nielsen, E.; Cheung, A.Y.; Ueda, T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol. 2008, 147, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.; Moore, I. The Arabidopsis Rab GTPase family: Another enigma variation. Curr. Opin. Plant Biol. 2002, 5, 518–528. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Angers, C.G.; Merz, A.J. New links between vesicle coats and Rab-mediated vesicle targeting. Semin. Cell Dev. Biol. 2011, 22, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Reddy, M.K.; Sopory, S.K.; Agarwal, P.K. Plant Rabs: Characterization, Functional Diversity, and Role in Stress Tolerance. Plant Mol. Biol. Rep. 2009, 27, 417–430. [Google Scholar] [CrossRef]
- Schwartz, S.L.; Cao, C.; Pylypenko, O.; Rak, A.; Wandinger-Ness, A. Rab GTPases at a glance. J. Cell Sci. 2007, 120, 3905–3910. [Google Scholar] [CrossRef] [PubMed]
- Karim, S.; Alezzawi, M.; Garcia-Petit, C.; Solymosi, K.; Khan, N.Z.; Lindquist, E.; Dahl, P.; Hohmann, S.; Aronsson, H. A novel chloroplast localized Rab GTPase protein CPRabA5e is involved in stress, development, thylakoid biogenesis and vesicle transport in Arabidopsis. Plant Mol. Biol. 2014, 84, 675–692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, T.; Yamaguchi, M.; Uchimiya, H.; Nakano, A. Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 2001, 20, 4730–4741. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Fujimoto, M.; Okatani, Y.; Nishiyama, T.; Goh, T.; Ito, E.; Dainobu, T.; Nishitani, A.; Uemura, T.; Sato, M.H.; et al. A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 2011, 13, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Miyakawa, N.; Fujimoto, M.; Uemura, T.; Nakano, A.; Ueda, T. Endosomal trafficking pathway regulated by ARA6, a RAB5 GTPase unique to plants. Small GTPases 2012, 3, 23–27. [Google Scholar] [CrossRef] [PubMed]
- Podell, S.; Gribskov, M. Predicting N-terminal myristoylation sites in plant proteins. BMC Genom. 2004, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishitani, M.; Liu, J.P.; Halfter, U.; Kim, C.S.; Shi, W.M.; Zhu, J.K. SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1677. [Google Scholar] [CrossRef] [PubMed]
- Boisson, B.; Meinnel, T. A continuous assay of myristoyl-CoA:protein N-myristoyltransferase for proteomic analysis. Anal. Biochem. 2003, 322, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Hoepflinger, M.C.; Geretschlaeger, A.; Sommer, A.; Hoeftberger, M.; Nishiyama, T.; Sakayama, H.; Hammerl, P.; Tenhaken, R.; Ueda, T.; Foissner, I. Molecular and biochemical analysis of the first ARA6 homologue, a RAB5 GTPase, from green algae. J. Exp. Bot. 2013, 64, 5553–5568. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Uchida, W.; Arakawa, S.; Ito, E.; Dainobu, T.; Ebine, K.; Takeuchi, M.; Sato, K.; Ueda, T.; Nakano, A. VPS9a, the common activator for two distinct types of Rab5 GTPases, is essential for the development of Arabidopsis thaliana. Plant Cell 2007, 19, 3504–3515. [Google Scholar] [CrossRef] [PubMed]
- Ebine, K.; Ueda, T. Unique mechanism of plant endocytic/vacuolar transport pathways. J. Plant Res. 2009, 122, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, T.; Nakano, A.; Ueda, T. The Plant-Specific RAB5 GTPase ARA6 is Required for Starch and Sugar Homeostasis in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Allu, A.D.; Soja, A.M.; Wu, A.; Szymanski, J.; Balazadeh, S. Salt stress and senescence: Identification of cross-talk regulatory components. J. Exp. Bot. 2014, 65, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Pitakrattananukool, S.; Sitthiphrom, S.; Cutler, R.W.; Anuntalabhochai, S. Molecular cloning of senescence related-cDNA-osrab7 from thai jasmine rice (Oryza sativa L. cv. KDML 105). Int. Res. J. Plant Sci. 2013, 4, 109–116. [Google Scholar]
- Zimmermann, P.; Hirsch-Hoffmann, M.; Hennig, L.; Gruissem, W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004, 136, 2621–2632. [Google Scholar] [CrossRef] [PubMed]
- Grennan, A.K. Genevestigator. Facilitating Web-based gene-expression analysis. Plant Physiol. 2006, 141, 1164–1166. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Lohman, K.N.G.S.; John, M.C.; Amasino, R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant. 1994, 92, 322–328. [Google Scholar] [CrossRef]
- Oh, S.A.; Lee, S.Y.; Chung, I.K.; Lee, C.H.; Nam, H.G. A senescence-associated gene of Arabidopsis thaliana is distinctively regulated during natural and artificially induced leaf senescence. Plant Mol. Biol. 1996, 30, 739–754. [Google Scholar] [CrossRef]
- Weaver, L.M.; Gan, S.S.; Quirino, B.; Amasino, R.M. A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 1998, 37, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Fischer-Kilbienski, I.; Miao, Y.; Roitsch, T.; Zschiesche, W.; Humbeck, K.; Krupinska, K. Nuclear targeted AtS40 modulates senescence associated gene expression in Arabidopsis thaliana during natural development and in darkness. Plant Mol. Biol. 2010, 73, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Schenk, P.M.; Kazan, K.; Rusu, A.G.; Manners, J.M.; Maclean, D.J. The SEN1 gene of Arabidopsis is regulated by signals that link plant defence responses and senescence. Plant Physiol. Biochem. 2005, 43, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Pourtau, N.J.R.; Pelzer, E.; Pallas, J.; Wingler, A. Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 2006, 224, 556–568. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Tsunoda, H.; Suzuki, Y.; Makino, A.; Ishida, H. RBCS1A and RBCS3B, two major members within the Arabidopsis RBCS multigene family, function to yield sufficient Rubisco content for leaf photosynthetic capacity. J. Exp. Bot. 2012, 63, 2159–2170. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M.; Tsunoda, H.; Suzuki, Y.; Makino, A.; Ishida, H. Circadian expression of the PpLhcb2 gene encoding a major light-harvesting chlorophyll a/b-binding protein in the moss Physcomitrella patens. Plant Cell Physiol. 2004, 45, 68–76. [Google Scholar]
- Johansson, O.N.; Fantozzi, E.; Fahlberg, P.; Nilsson, A.K.; Buhot, N.; Tör, M.; Andersson, M.X. Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J. 2014, 79, 466–476. [Google Scholar] [CrossRef] [PubMed]
- Julkowska, M.M.; Testerink, C. Tuning plant signaling and growth to survive salt. Trends Plant Sci. 2015, 20, 586–594. [Google Scholar] [CrossRef] [PubMed]
- Garcia, C.; Khan, N.Z.; Nannmark, U.; Aronsson, H. The chloroplast protein CPSAR1, dually localized in the stroma and the inner envelope membrane, is involved in thylakoid biogenesis. Plant J. 2010, 63, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, H.; Jarvis, P. A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett. 2002, 529, 215–220. [Google Scholar] [CrossRef]
- Karimi, M.; Inze, D.; Depicker, A. GATEWAY((TM)) vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Koncz, C.; Nemeth, K.; Redei, G.P.; Schell, J. T-DNA Insertional Mutagenesis in Arabidopsis. Plant Mol. Biol. 1992, 20, 963–976. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.; Thompson, W.; Kriedemann, P. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Aronsson, H.; Combe, J.; Jarvis, P. Unusual nucleotide-binding properties of the chloroplast protein import receptor, atToc33. FEBS Lett. 2003, 544, 79–85. [Google Scholar] [CrossRef]
- Yamatani, H.; Sato, Y.; Masuda, Y.; Kato, Y.; Morita, R.; Fukunaga, K.; Nagamura, Y.; Nishimura, M.; Sakamoto, W.; Tanaka, A.; et al. NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll—Protein complexes during leaf senescence. Plant J. 2013, 74, 652–662. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, C.; Karim, S.; Zhang, H.; Aronsson, H. Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS). Int. J. Mol. Sci. 2017, 18, 309. https://doi.org/10.3390/ijms18020309
Yin C, Karim S, Zhang H, Aronsson H. Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS). International Journal of Molecular Sciences. 2017; 18(2):309. https://doi.org/10.3390/ijms18020309
Chicago/Turabian StyleYin, Congfei, Sazzad Karim, Hongsheng Zhang, and Henrik Aronsson. 2017. "Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS)" International Journal of Molecular Sciences 18, no. 2: 309. https://doi.org/10.3390/ijms18020309




