Genetic Variation in Choline-Metabolizing Enzymes Alters Choline Metabolism in Young Women Consuming Choline Intakes Meeting Current Recommendations
Abstract
:1. Introduction
2. Results
2.1. Genotype Distribution
2.2. CHKA rs10791957
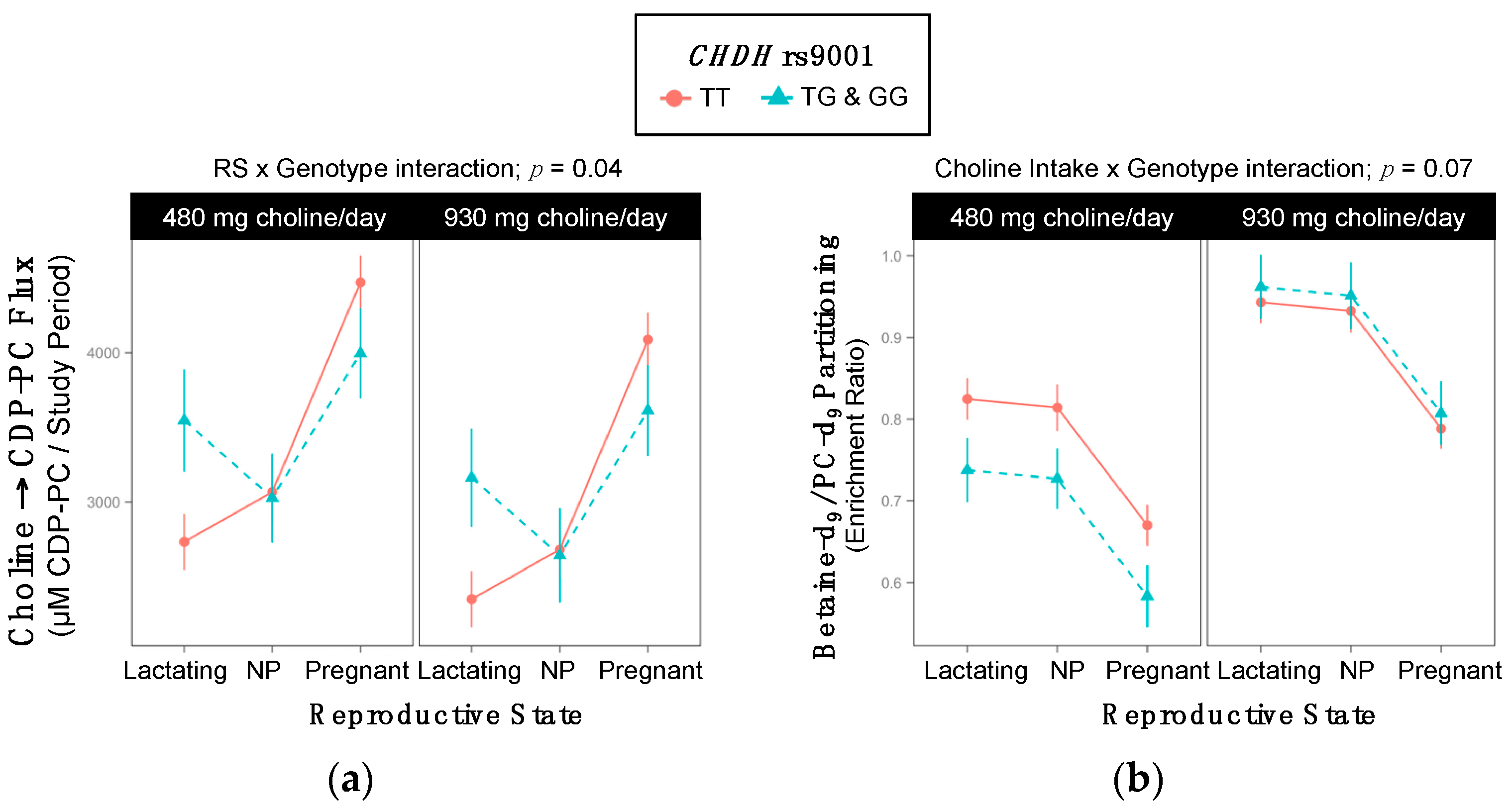
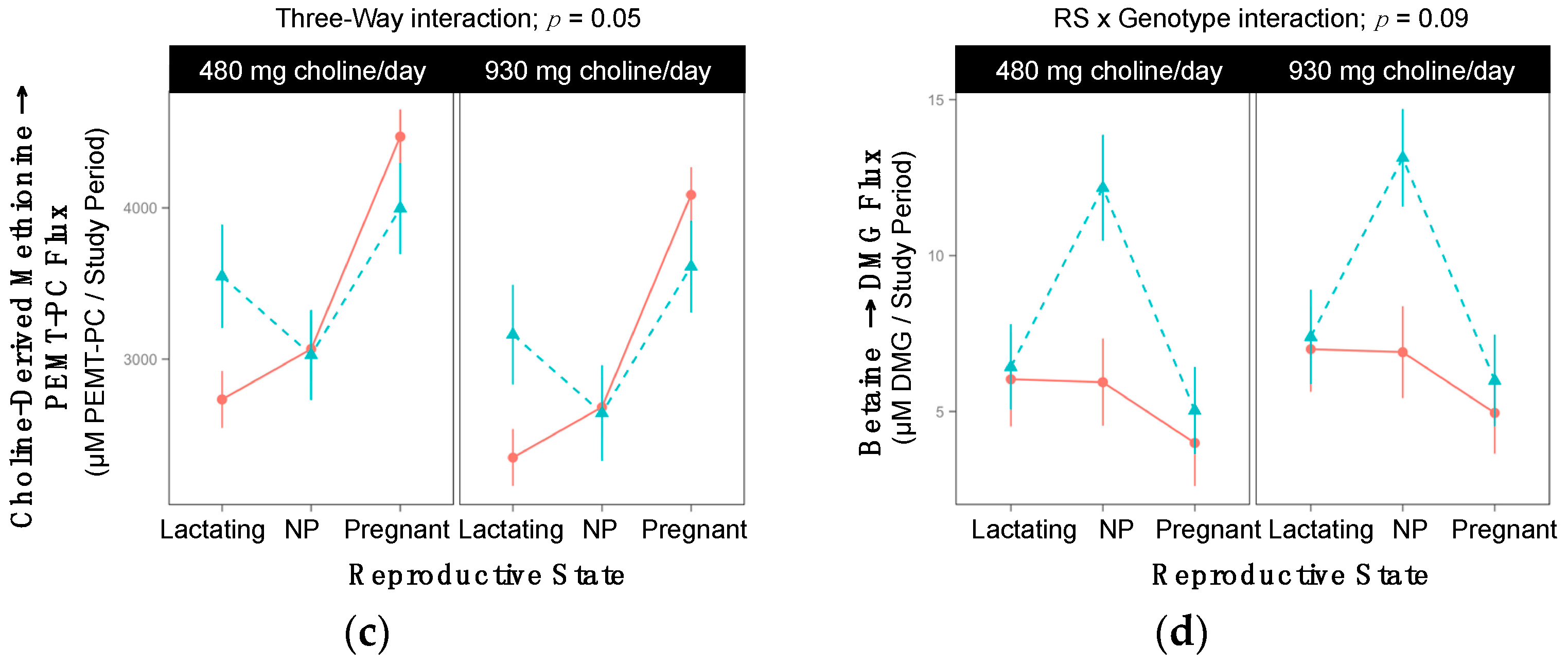
2.3. CHDH rs9001
2.4. CHDH rs12676
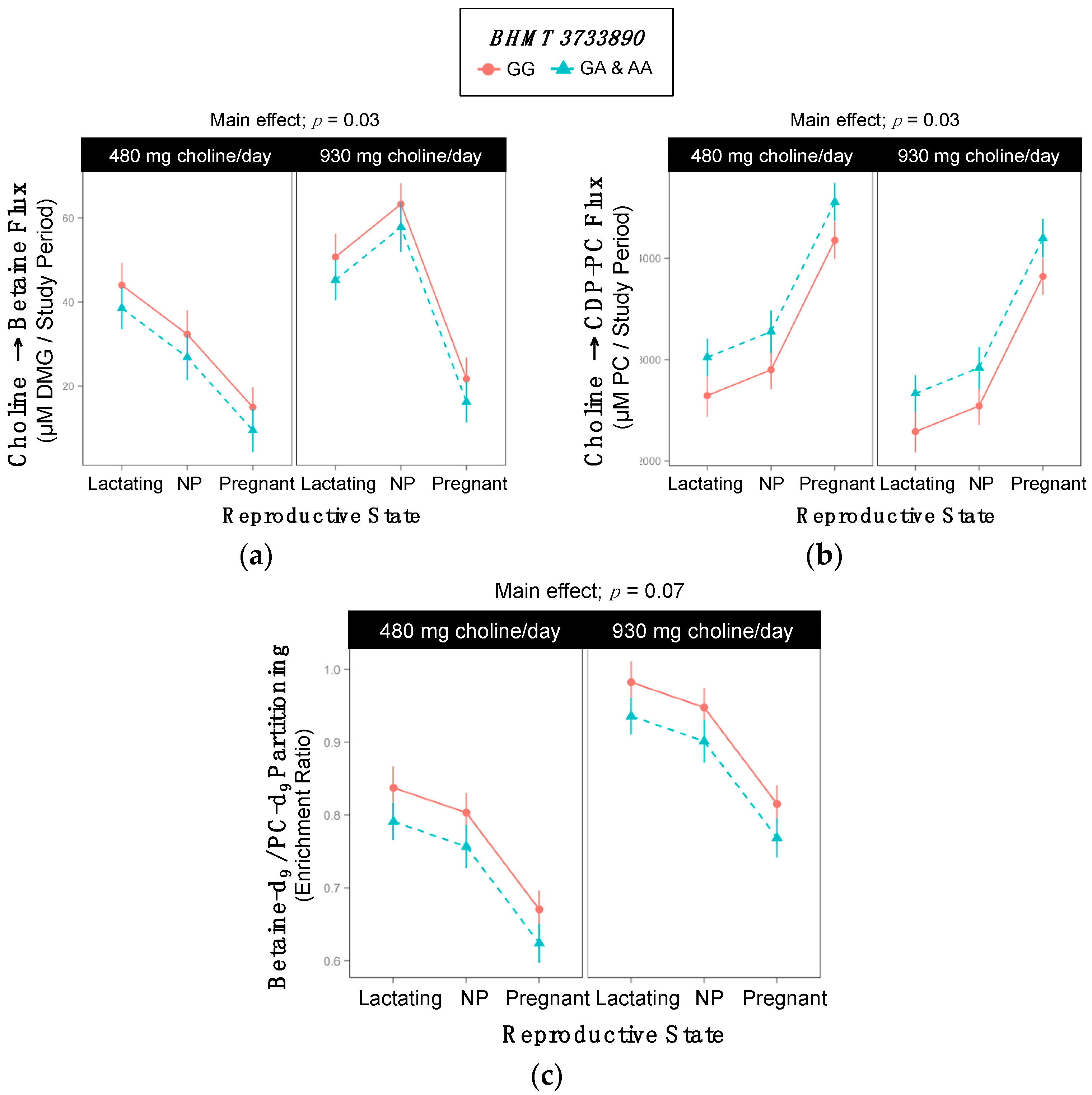
2.5. BHMT rs3733890
2.6. PEMT rs4646343
2.7. PEMT rs7946
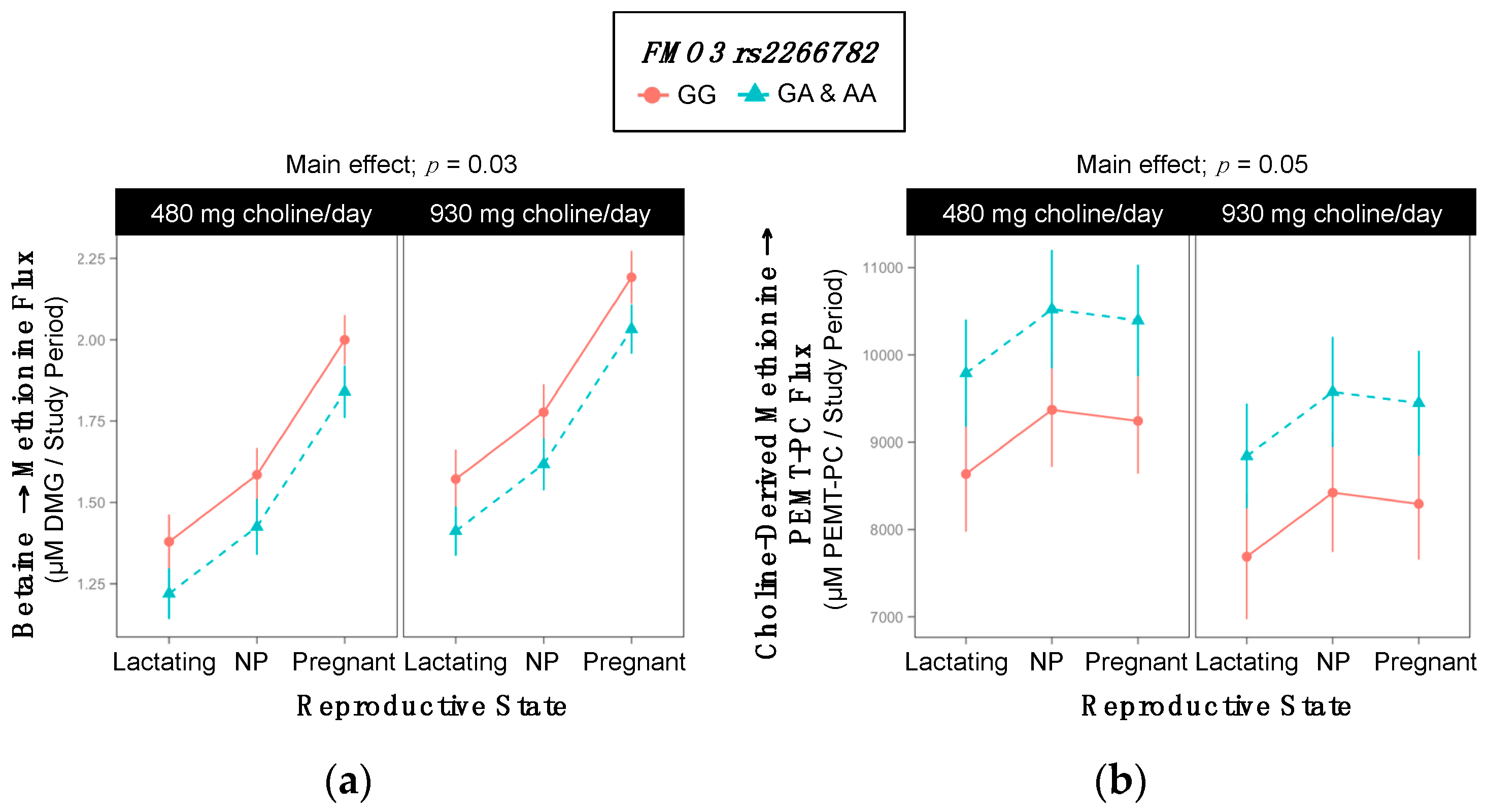
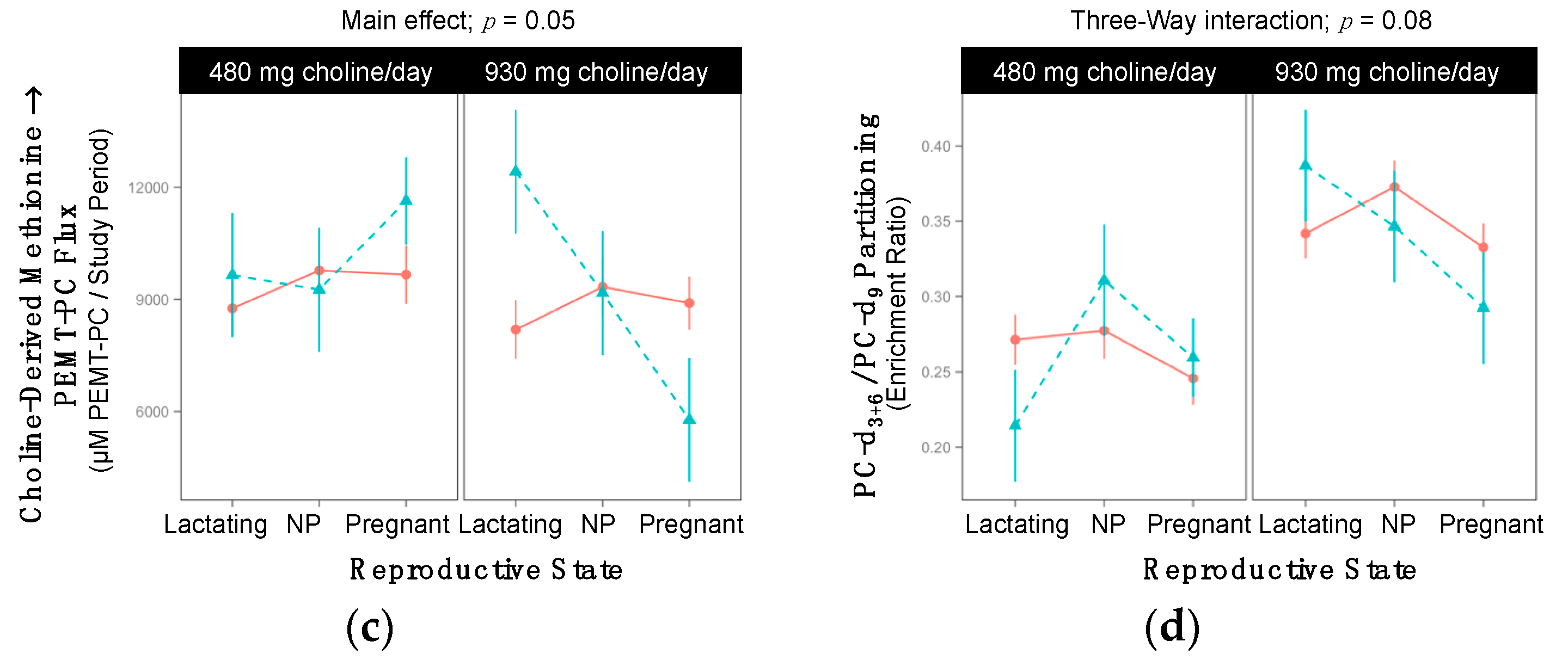
2.8. FMO3 rs2266782
2.9. SLC44A1 rs7873937
2.10. SLC44A1 rs3199966
3. Discussion
3.1. CHKA (dbSNP: rs10791957; NC_000011.10: g.68100081 C > A)
3.2. CHDH (dbSNP: rs9001; c.119 A > C; p.Glu40Ala) and (dbSNP: rs12676; c.233T > G; p.Leu78Arg)
3.3. BHMT (dbSNP: rs3733890; c.716 G > A, also Known as c.742 G > A; p.Arg239Gln)
3.4. PEMT (dbSNP: rs4646343; c.2768 C > A REV) and (dbSNP: rs7946; c.5465 G > A REV; p.Val175Met)
3.5. FMO3 (dbSNP: rs2266782; c.472 G > A; p.Glu158Lys)
3.6. SLC44A1 (dbSNP: rs7873937; NC_000009.11:g.108089321 G > C) and SLC44A1 (dbSNP: rs3199966; c.1930 T > G; p.Ser644Ala)
3.7. Study Limitations
3.8. Conclusions
4. Materials and Methods
4.1. Participants and Study Design
4.2. Genotyping
4.3. Enrichment of Choline Metabolites
4.4. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AI | Adequate intake |
| BHMT | Betaine homocysteine methyltransferase |
| CDP | Cytidine diphosphate |
| CHKA | Choline kinase-α |
| CHDH | Choline dehydrogenase |
| CI | Confidence interval |
| DMG | Dimethylglycine |
| FMO3 | Flavin monooxygenase isoform 3 |
| GC-MS | Gas chromatography-mass spectrometry |
| NTD | Neural tube defect |
| PE | Phosphatidylethanolamine |
| PEMT | Phosphatidylethanolamine N-methyltransferase |
| PC | Phosphatidylcholine |
| REV | SNP is identified on the reverse strand |
| RS | Reproductive state |
| SAH | S-adenosylhomocysteine |
| SAM | S-adenosylmethionine |
| SLC44A1 | Solute carrier 44A1 |
| MTHFR | Methylene tetrahydrofolate reductase |
| NAFLD | Non-alcoholic fatty liver disease |
| NP | Non-pregnant |
| WT | Wildtype |
References
- Caudill, M.A.; Miller, J.W.; III, J.F.G. Folate, choline, vitamin B 12, and vitamin B 6. In Stipanuk MH and Caudill MA. Biochemical, Physiological and Molecular Aspects of Human Nutrition; Elsevier Saunders: St. Louis, MO, USA, 2012; pp. 565–608. [Google Scholar]
- Li, Z.; Vance, D.E. Phosphatidylcholine and choline homeostasis. J. Lipid Res. 2008, 49, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Delong, C.J.; Shen, Y.; Michael, J.; Cui, Z. Molecular Distinction of Phosphatidylcholine Synthesis between the CDP-Choline Pathway and Pathway Molecular Distinction of Phosphatidylcholine Synthesis between the CDP-Choline Pathway and Phosphatidy. J. Biol. Chem. 1999, 274, 29683–29688. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US) Standing Committee on the Scientific Evaluation of Dietary Reference Intakes and its Panel on Folate, Other B Vitamins, and C. Dietary Reference Intakes for Thiamin, Riboflavin, Niacin, Vitamin B6, Folate, Vitamin B12, Pantothenic Acid, Biotin, and Choline; National Academies Press (US): Washington, DC, USA, 1998. [Google Scholar]
- Yan, J.; Wang, W.; Iii, J.F.G.; Malysheva, O.; Brenna, J.T.; Stabler, S.P.; Allen, R.H.; Caudill, M.A. MTHFR C677T genotype influences the isotopic enrichment of one-carbon metabolites in folate-compromised men consuming. Am. J. Clin. Nutr. 2011, 93, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Jiang, X.; West, A.A.; Perry, C.A.; Malysheva, O.V.; Devapatla, S.; Pressman, E.; Vermeylen, F.; Stabler, S.P.; Allen, R.H.; et al. Maternal choline intake modulates maternal and fetal biomarkers of choline metabolism in humans. Am. J. Clin. Nutr. 2012, 95, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Fischer, L.M.; DaCosta, K.A.; Kwock, L.; Stewart, P.W.; Lu, T.S.; Stabler, S.P.; Allen, R.H.; Zeisel, S.H. Sex and menopausal status influence human dietary requirements for the nutrient choline. Am. J. Clin. Nutr. 2007, 85, 1275–1285. [Google Scholar] [PubMed]
- Da Costa, K.-A.; Kozyreva, O.G.; Song, J.; Galanko, J.A.; Fischer, L.M.; Zeisel, S.H. Common genetic polymorphisms affect the human requirement for the nutrient choline. FASEB J. 2006, 20, 1336–1344. [Google Scholar] [CrossRef] [PubMed]
- Ganz, A.B.; Shields, K.; Fomin, V.G.; Lopez, Y.S.; Mohan, S.; Lovesky, J.; Chuang, J.C.; Ganti, A.; Carrier, B.; Yan, J.; et al. Genetic impairments in folate enzymes increase dependence on dietary choline for phosphatidylcholine production at the expense of betaine synthesis. FASEB J. 2016, 30, 3321–3333. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, K.A.; Corbin, K.D.; Niculescu, M.D.; Galanko, J.A.; Zeisel, S.H. Identification of new genetic polymorphisms that alter the dietary requirement for choline and vary in their distribution across ethnic and racial groups. FASEB J. 2014, 28, 2970–2978. [Google Scholar] [CrossRef] [PubMed]
- Jeff, J.M.; Armstrong, L.L.; Ritchie, M.D.; Denny, J.C.; Kho, A.N.; Basford, M.A.; Wolf, W.A.; Pacheco, J.A.; Li, R.; Chisholm, R.L.; et al. Admixture mapping and subsequent fine-mapping suggests a biologically relevant and novel association on chromosome 11 for type 2 diabetes in African Americans. PLoS ONE 2014, 9, e86931. [Google Scholar] [CrossRef] [PubMed]
- Schläwicke Engström, K.; Nermell, B.; Concha, G.; Strömberg, U.; Vahter, M.; Broberg, K. Arsenic metabolism is influenced by polymorphisms in genes involved in one-carbon metabolism and reduction reactions. Mutat. Res. 2009, 667, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Zeisel, S.H.; Lee, Y.L.; Wetmur, J.G.; Teitelbaum, S.L.; Bradshaw, P.T.; Neugut, A.I.; Santella, R.M.; Chen, J. Choline metabolism and risk of breast cancer in a population-based study. FASEB J. 2008, 22, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.R.; Lao, S.; Wang, T.; Galanko, J.A.; Zeisel, S.H. Choline dehydrogenase polymorphism rs12676 is a functional variation and is associated with changes in human sperm cell function. PLoS ONE 2012, 7, e36047. [Google Scholar] [CrossRef] [PubMed]
- Mostowska, A.; Hozyasz, K.K.; Wojcicki, P.; Dziegelewska, M.; Jagodzinski, P.P. Associations of folate and choline metabolism gene polymorphisms with orofacial clefts. J. Med. Genet. 2010, 47, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Gammon, M.D.; Wetmur, J.G.; Bradshaw, P.T.; Susan, L.; Neugut, A.I.; Santella, R.M.; Chen, J. NIH B-vitamin Intake, One-carbon Metabolism and Survival among a Population-based Study of Women with Breast Cancer. Biomarkers 2009, 17, 2109–2116. [Google Scholar] [CrossRef] [PubMed]
- Shaw, G.M.; Lu, W.; Zhu, H.; Yang, W.; Briggs, F.B.S.; Carmichael, S.L.; Barcellos, L.F.; Lammer, E.J.; Finnell, R.H. 118 SNPs of folate-related genes and risks of spina bifida and conotruncal heart defects. BMC Med. Genet. 2009, 10, 49. [Google Scholar] [CrossRef] [PubMed]
- Boyles, A.L.; Billups, A.V.; Deak, K.L.; Siegel, D.G.; Mehltretter, L.; Slifer, S.H.; Bassuk, A.G.; Kessler, J.A.; Reed, M.C.; Nijhout, H.F.; et al. Neural tube defects and folate pathway genes: Family-based association tests of gene-gene and gene-environment interactions. Environ. Health Perspect. 2006, 114, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Langberg, K.A.; Mondal, A.K.; Das, S.K. Phospholipid biosynthesis genes and susceptibility to obesity: Analysis of expression and polymorphisms. PLoS ONE 2013, 8, e65303. [Google Scholar] [CrossRef]
- Song, J.; da Costa, K.A.; Fischer, L.M.; Kohlmeier, M.; Kwock, L.; Wang, S.; Zeisel, S.H. Polymorphism of the PEMT gene and susceptibility to nonalcoholic fatty liver disease (NAFLD). FASEB J. 2005, 19, 1266–1271. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H. People with fatty liver are more likely to have the PEMT rs7946 SNP, yet populations with the mutant allele do not have fatty liver. FASEB J. 2006, 20, 2181–2182. [Google Scholar] [CrossRef]
- Phillips, I.R.; Shephard, E.A. Primary Trimethylaminuria; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Gibellini, F.; Smith, T.K. The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 2010, 62, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Trousil, S.; Lee, P.; Pinato, D.J.; Ellis, J.; Dina, R.; Aboagye, E.O.; Keun, H.C.; Sharma, R. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase α overexpression and a hyperactivated deacylation pathway. Cancer Res. 2014, 74, 6867–6878. [Google Scholar] [CrossRef] [PubMed]
- Ramírez de Molina, A.; Gallego-Ortega, D.; Sarmentero-Estrada, J.; Lagares, D.; Gómez del Pulgar, T.; Bandrés, E.; García-Foncillas, J.; Lacal, J.C. Choline kinase as a link connecting phospholipid metabolism and cell cycle regulation: Implications in cancer therapy. Int. J. Biochem. Cell Biol. 2008, 40, 1753–1763. [Google Scholar] [CrossRef] [PubMed]
- Gréchez-Cassiau, A.; Feillet, C.; Guérin, S.; Delaunay, F. The hepatic circadian clock regulates the choline kinase α gene through the BMAL1-REV-ERB α axis. Chronobiol. Int. 2015, 32, 774–784. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, R.L.; Zhao, Y.; Koonen, D.P.Y.; Sletten, T.; Su, B.; Lingrell, S.; Cao, G.; Peake, D.A.; Kuo, M.-S.; Proctor, S.D.; et al. Impaired de novo choline synthesis explains why phosphatidylethanolamine N-methyltransferase-deficient mice are protected from diet-induced obesity. J. Biol. Chem. 2010, 285, 22403–22413. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H. Gene response elements, genetic polymorphisms and epigenetics influence the human dietary requirement for choline. IUBMB Life 2007, 59, 380–387. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Dahiya, S.; Provencher, H.; Muir, B.; Carney, E.; Coser, K.; Shioda, T.; Ma, X.-J.; Sgroi, D.C. The Prognostic Biomarkers HOXB13, IL17BR, and CHDH Are Regulated by Estrogen in Breast Cancer. Clin. Cancer Res. 2007, 13, 6327–6334. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Feng, Q.; Lee, C.; Wang, S.; Pelleymounter, L.L.; Moon, I.; Eckloff, B.W.; Wieben, E.D.; Schaid, D.J.; Yee, V.; et al. Human betaine-homocysteine methyltransferase (BHMT) and BHMT2: Common gene sequence variation and functional characterization. Mol. Genet. Metab. 2008, 94, 326–335. [Google Scholar] [CrossRef] [PubMed]
- Morin, I.; Platt, R.; Weisberg, I.; Sabbaghian, N.; Wu, Q.; Garrow, T.A.; Rozen, R. Common variant in betaine-homocysteine methyltransferase (BHMT) and risk for spina bifida. Am. J. Med. Genet. A 2003, 119A, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, N.D.; Vance, D.E. Kinetic mechanism of phosphatidylethanolamine N-methyltransferase. J. Biol. Chem. 1988, 263, 16864–16871. [Google Scholar] [PubMed]
- Resseguie, M.E.; da Costa, K.-A.; Galanko, J.A.; Patel, M.; Davis, I.J.; Zeisel, S.H. Aberrant estrogen regulation of PEMT results in choline deficiency-associated liver dysfunction. J. Biol. Chem. 2011, 286, 1649–1658. [Google Scholar] [CrossRef] [PubMed]
- Pynn, C.J.; Henderson, N.G.; Clark, H.; Koster, G.; Bernhard, W.; Postle, A.D. Specificity and rate of human and mouse liver and plasma phosphatidylcholine synthesis analyzed in vivo. J. Lipid Res. 2011, 52, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T.C.; Fulgoni, V.L. Assessment of Total Choline Intakes in the United States. J. Am. Coll. Nutr. 2016, 35, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Craciun, S.; Balskus, E.P. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc. Natl. Acad. Sci. USA 2012, 109, 21307–21312. [Google Scholar] [CrossRef] [PubMed]
- Koukouritaki, S.B.; Poch, M.T.; Cabacungan, E.T.; McCarver, D.G.; Hines, R.N. Discovery of novel flavin-containing monooxygenase 3 (FMO3) single nucleotide polymorphisms and functional analysis of upstream haplotype variants. Mol. Pharmacol. 2005, 68, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Ling, A.V.; Manthena, P.V.; Gearing, M.E.; Graham, M.J.; Crooke, R.M.; Croce, K.J.; Esquejo, R.M.; Clish, C.B.; Torrecilla, E.; et al. Flavin-containing monooxygenase 3 as a potential player in diabetes-associated atherosclerosis. Nat. Commun. 2015, 6, 6498. [Google Scholar] [CrossRef] [PubMed]
- Bennett, B.J.; Vallim, T.Q.A.; Wang, Z.; Shih, D.M.; Meng, Y.; Gregory, J.; Allayee, H.; Lee, R.; Graham, M.; Crooke, R.; et al. Trimethylamine-N-Oxide, a Metabolite Associated with Atherosclerosis, Exhibits Complex Genetic and Dietary Regulation. Changes 2012, 29, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.H.W.; Wang, Z.; Levison, B.S.; Koeth, R.A.; Britt, E.B.; Fu, X.; Wu, Y.; Hazen, S.L. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013, 368, 1575–1584. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Warrier, M.; Shih, D.M.; Burrows, A.C.; Ferguson, D.; Gromovsky, A.D.; Brown, A.L.; Marshall, S.; McDaniel, A.; Schugar, R.C.; Wang, Z.; et al. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. 2015, 10, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.E.; Taesuwan, S.; Malysheva, O.V.; Bender, E.; Yan, J.; Caudill, M.A. Choline and one-carbon metabolite response to egg, beef and fish among healthy young men: A short-term randomized clinical study. Clin. Nutr. Exp. 2016, 10, 1–11. [Google Scholar] [CrossRef]
- PB|CTL1–5 (Plasma Membrane). Available online: http://www.reactome.org/PathwayBrowser/#/R-HSA-425366&SEL=R-HSA-444452 (accessed on 14 December 2016).
- SLC44A1; Choline Transporter-Like Protein 1. Available online: https://www.nextprot.org/entry/NX_Q8WWI5/ (accessed on 14 December 2016).
- Traiffort, E.; O’Regan, S.; Ruat, M. The choline transporter-like family SLC44: Properties and roles in human diseases. Mol. Asp. Med. 2013, 34, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Michel, V.; Singh, R.K.; Bakovic, M. The impact of choline availability on muscle lipid metabolism. Food Funct. 2011, 2, 53–62. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing, 2014; R Foundation for Statistical Computing: Vienna, Austria, 2016. [Google Scholar]
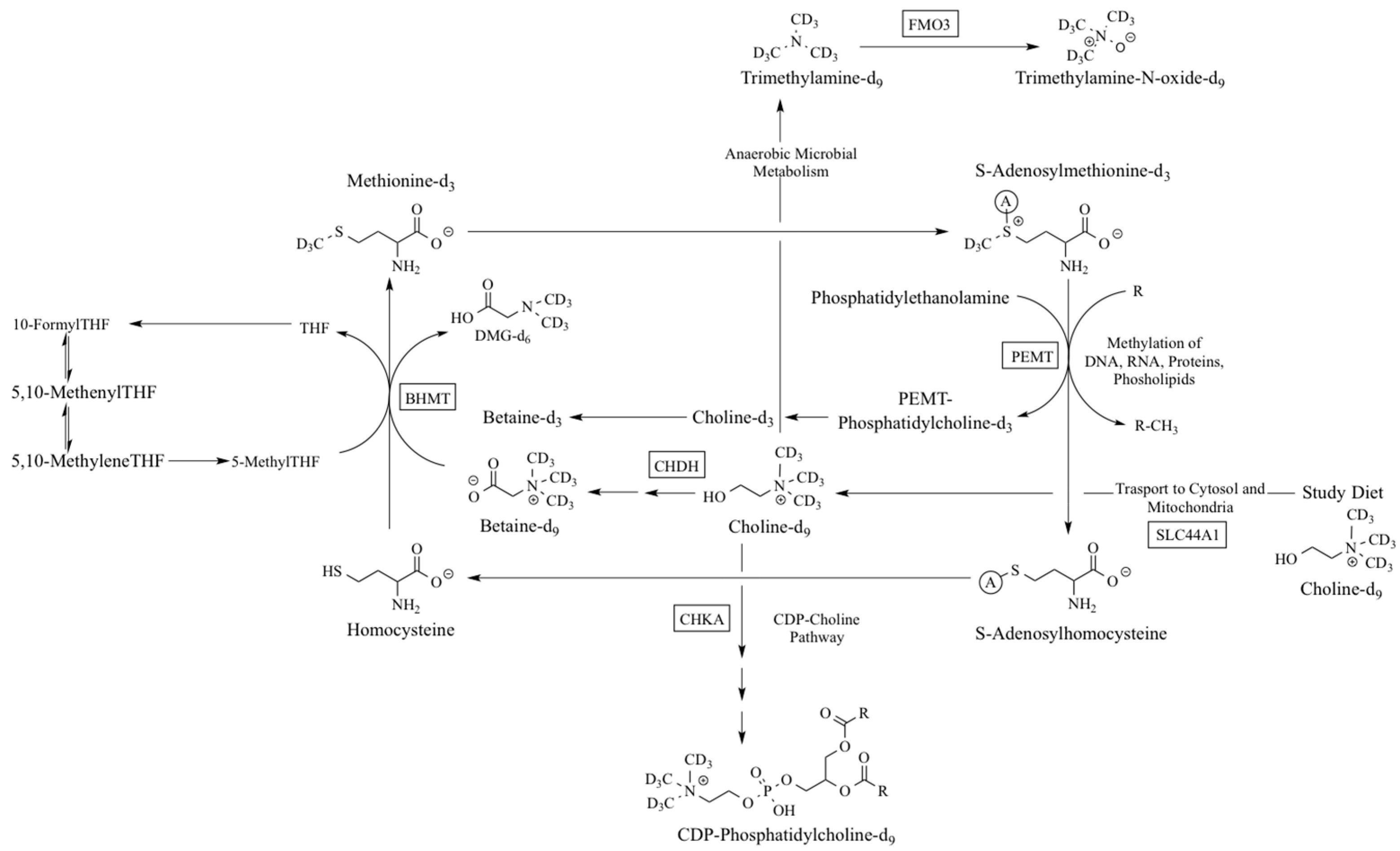






| Gene | Function | SNP | Choline Deficiency Risk | Disease Associations | References |
|---|---|---|---|---|---|
| CHKA | Phosphorylates choline, first step in CDP-choline pathway | rs10791957 | ↓ risk organ dysfunction * | ↓ risk type 2 diabetes | [10,11] |
| CHDH | First step in oxidation of choline to betaine | rs9001 | ↓ risk organ dysfunction | ↑ arsenic methylation | [10,12] |
| rs12676 | ↑ risk organ dysfunction ** | ↑ breast cancer risk | [13,14] | ||
| ↓ sperm ATP and altered sperm motility | |||||
| BHMT | Converts homocysteine to methionine using betaine as a methyl donor | rs3733890 | - | ↓ breast cancer mortality | [10,15,16,17,18] |
| ↑ orofacial cleft | |||||
| ↑ spina bifida (mixed results) | |||||
| PEMT | Uses SAM to triply methylate PE to form PC (endogenous choline synthesis) | rs4646343 | ↑ risk organ dysfunction | ↑ PEMT expression in adipose | [19] |
| ↑ waist to hip ratio | |||||
| rs7946 | - | ↑ Non-alcoholic fatty liver disease | [20,21] | ||
| FMO3 | Converts TMA to TMAO | rs2266782 | - | ↑ trimethylaminuria | [22] |
| SLC44A1 | Transports choline across the cellular and mitochondrial membranes | rs7873937 | ↑ risk muscle damage | [10] | |
| rs3199966 | ↑ risk muscle damage | [10] |
| Group | 480 mg Choline/day | 930 mg Choline/day | ||||
|---|---|---|---|---|---|---|
| # Of Variant Alleles | 0 | 1 | 2 | 0 | 1 | 2 |
| CHKA rs10791957 | ||||||
| Lactating | 2 | 5 | 6 | 2 | 4 | 7 |
| Non-pregnant | 2 | 5 | 3 | 2 | 7 | 2 |
| Pregnant | 2 | 6 | 5 | 1 | 3 | 9 |
| CHDH rs9001 | ||||||
| Lactating | 11 | 1 | 1 | 10 | 2 | 1 |
| Non-pregnant | 6 | 4 | 0 | 9 | 2 | 0 |
| Pregnant | 10 | 2 | 1 | 10 | 2 | 1 |
| CHDH rs12676 | ||||||
| Lactating | 5 | 7 | 1 | 9 | 3 | 1 |
| Non-pregnant | 7 | 1 | 2 | 5 | 6 | 0 |
| Pregnant | 6 | 6 | 1 | 8 | 4 | 1 |
| BHMT rs3733890 | ||||||
| Lactating | 5 | 6 | 2 | 3 | 8 | 2 |
| Non-pregnant | 4 | 6 | 0 | 9 | 1 | 1 |
| Pregnant | 8 | 4 | 1 | 6 | 6 | 1 |
| PEMT rs4646343 | ||||||
| Lactating | 6 | 4 | 3 | 6 | 6 | 1 |
| Non-pregnant | 5 | 5 | 0 | 6 | 4 | 1 |
| Pregnant | 5 | 5 | 3 | 6 | 4 | 3 |
| PEMT rs7946 | ||||||
| Lactating | 2 | 2 | 9 | 2 | 2 | 9 |
| Non-pregnant | 2 | 5 | 3 | 3 | 5 | 3 |
| Pregnant | 0 | 6 | 7 | 2 | 3 | 8 |
| FMO3 rs2266782 | ||||||
| Lactating | 7 | 5 | 1 | 3 | 8 | 2 |
| Non-pregnant | 6 | 3 | 1 | 4 | 4 | 3 |
| Pregnant | 6 | 4 | 3 | 7 | 4 | 1 |
| SLC44A1 rs7873937 | ||||||
| Lactating | 11 | 2 | 0 | 11 | 2 | 0 |
| Non-pregnant | 8 | 2 | 0 | 9 | 2 | 0 |
| Pregnant | 9 | 3 | 1 | 11 | 2 | 0 |
| SLC44A1 rs3199966 | ||||||
| Lactating | 10 | 3 | 0 | 11 | 2 | 0 |
| Non-pregnant | 8 | 2 | 0 | 8 | 3 | 0 |
| Pregnant | 8 | 4 | 1 | 9 | 4 | 0 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| Choline-Derived Methionine → PEMT-PC | 11,512 ±664 | 8840 ±291 | 0.0005 |
| PC-d3+6/PC-d9 | 0.33 ±0.015 | 0.30 ±0.007 | 0.09 |
| Metabolic Outcome and Group | WT | Variant | p-Value |
|---|---|---|---|
| Choline → CDP-PC | |||
| RS × Gene Interaction; p = 0.04 | |||
| Lactating | 2542 ±167 | 3355 ±324 | 0.09 |
| Non-pregnant | 2875 ±185 | 2836 ±295 | >0.99 |
| Pregnant | 4279 ±159 | 3804 ±290 | 0.5 |
| Metabolic Outcome and Group | WT | Variant | p-Value |
|---|---|---|---|
| Betaine-d9/PC-d9 | |||
| Cho × Gene Interaction; p = 0.07 | |||
| 480 mg Choline/day | 0.77 ±0.02 | 0.68 ±0.03 | 0.06 |
| 930 mg Choline/day | 0.89 ±0.02 | 0.91 ±0.04 | >0.99 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| PC-d3+6/PC-d9 | 0.30 ±0.008 | 0.32 ±0.009 | 0.055 |
| Metabolic Outcome | 480 mg Choline/day WT | 480 mg Choline/day Variant | p-Value | 930 mg Choline/day WT | 930 mg Choline/day Variant | p-Value |
|---|---|---|---|---|---|---|
| Choline → CDP-PC 3-Way Interaction; p = 0.05 | ||||||
| Lactating | 3538 ±364 | 2345 ±264 | 0.06 | 2779 ±246 | 2394 ±350 | >0.99 |
| Non-pregnant | 3127 ±266 | 2782 ±403 | >0.99 | 2621 ±312 | 2764 ±285 | >0.99 |
| Pregnant | 4080 ±285 | 4809 ±263 | 0.39 | 4021 ±248 | 3609 ±313 | >0.99 |
| Choline-Derived Methionine → PEMT-PC 3-Way Interaction; p = 0.05 | ||||||
| Lactating | 12358 ±1130 | 7182 ±799 | 0.003 | 9151 ±799 | 8467 ±1304 | >0.99 |
| Non-pregnant | 9334 ±854 | 10451 ±1304 | >0.99 | 9338 ±1010 | 9272 ±922 | >0.99 |
| Pregnant | 9668 ±922 | 10782 ±854 | >0.99 | 8728 ±799 | 7929 ±1010 | >0.99 |
| Metabolic Outcome and Group | WT | Variant | p-Value |
|---|---|---|---|
| Betaine → DMG RS × Gene Interaction; p = 0.09 | |||
| Lactating | 6.5 ±1.3 | 6.9 ±1.3 | >0.99 |
| Non-pregnant | 6.4 ±1.3 | 12.7 ±1.5 | 0.01 |
| Pregnant | 4.5 ±1.2 | 5.5 ±1.3 | >0.99 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| Choline → Betaine | 38 ±3 | 32 ±3 | 0.2 |
| Choline → CDP-PC | 3063 ±122 | 3440 ±122 | 0.03 |
| Betaine-d9/PC-d9 | 0.82 ±0.02 | 0.77 ±0.02 | 0.07 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| PC-d3+6/PC-d9 | 0.32 ±0.009 | 0.30 ±0.008 | 0.05 |
| Metabolic Outcome and Group | WT | Variant | p-Value |
|---|---|---|---|
| PC-d3+6/PC-d9 RS × Gene Interaction; p = 0.09 | |||
| Lactating | 0.37 ±0.03 | 0.29 ±0.01 | 0.03 |
| Non-pregnant | 0.32 ±0.02 | 0.33 ±0.01 | >0.99 |
| Pregnant | 0.30 ±0.04 | 0.29 ±0.01 | >0.99 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| Choline-Derived Methionine → PEMT-PC | 7942 ±765 | 9481 ±312 | 0.07 |
| Metabolic Outcome | WT | Variant | p-Value |
|---|---|---|---|
| Betaine → Methionine | 1.8 ±0.05 | 1.6 ±0.05 | 0.03 |
| Choline-Derived Methionine → PEMT-PC | 8609 ±433 | 9761 ±384 | 0.05 |
| Metabolic Outcome and Group | 480 mg Choline/day | 930 mg Choline/day | p-Value |
|---|---|---|---|
| Betaine → Methionine Cho × Gene Interaction; p = 0.06 | |||
| WT | 1.60 ±0.06 | 1.70 ±0.06 | 0.9 |
| Variant | 1.51 ±0.12 | 1.95 ±0.11 | 0.03 |
| Metabolic Outcome and Group | 480 mg Choline/day WT | 480 mg Choline/day Variant | p-Value | 930 mg Choline/day WT | 930 mg Choline/day Variant | p-Value |
|---|---|---|---|---|---|---|
| Betaine → DMG 3-Way Interaction; p = 0.04 | ||||||
| Lactating | 6.2 ±1.5 | 6.7 ±3.3 | >0.99 | 7.4 ±1.5 | 5.7 ±3.3 | >0.99 |
| Non-pregnant | 9.2 ±1.6 | 5.6 ±3.3 | >0.99 | 7.5 ±1.5 | 19.6 ±3.3 | 0.02 |
| Pregnant | 3.8 ±1.5 | 4.5 ±2.3 | >0.99 | 5.6 ± 1.4 | 7.1 ±3.3 | >0.99 |
| Metabolic Outcome and Group | 480 mg Choline/day WT | 930 mg Choline/day WT | p-Value | 480 mg Choline/day Variant | 930 mg Choline/day Variant | p-Value |
|---|---|---|---|---|---|---|
| Betaine → DMG 3-Way Interaction; p = 0.04 | ||||||
| Lactating | 6.2 ±1.5 | 7.4 ±1.5 | >0.99 | 6.7 ±3.3 | 5.7 ±3.3 | >0.99 |
| Non-pregnant | 9.2 ±1.6 | 7.5 ±1.5 | >0.99 | 5.6 ±3.3 | 19.6 ±3.3 | 0.05 |
| Pregnant | 3.8 ±1.5 | 5.6 ±1.4 | >0.99 | 4.5 ±2.3 | 7.1 ±3.3 | >0.99 |
| Choline-Derived Methionine → PEMT-PC 3-Way Interaction; p = 0.05 | ||||||
| Lactating | 8759 ±741 | 8197 ±782 | >0.99 | 9648 ±1658 | 12417 ±1658 | >0.99 |
| Non-pregnant | 9772 ±829 | 9331 ±782 | >0.99 | 9257 ±1658 | 9172 ±1658 | >0.99 |
| Pregnant | 9660 ±782 | 8900 ±707 | >0.99 | 11635 ±1172 | 5780 ±1658 | 0.07 |
| PC-d3+6/PC-d9 3-Way Interaction; p = 0.08 | ||||||
| Lactating | 0.27 ±0.02 | 0.34 ±0.02 | 0.05 | 0.21 ±0.04 | 0.39 ± 0.04 | 0.02 |
| Non-pregnant | 0.28 ±0.02 | 0.37 ±0.02 | 0.005 | 0.31 ±0.04 | 0.35 ±0.04 | >0.99 |
| Pregnant | 0.25 ±0.02 | 0.33 ±0.02 | 0.006 | 0.26 ±0.03 | 0.29 ±0.04 | >0.99 |
| Metabolic Outcome and Group | 480 mg Choline/day | 930 mg Choline/day | p-Value |
|---|---|---|---|
| Betaine → Methionine Cho × Gene Interaction; p = 0.08 | |||
| WT | 1.60 ±0.06 | 1.70 ±0.06 | >0.99 |
| Variant | 1.52 ±0.10 | 1.90 ±0.11 | 0.04 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ganz, A.B.; Cohen, V.V.; Swersky, C.C.; Stover, J.; Vitiello, G.A.; Lovesky, J.; Chuang, J.C.; Shields, K.; Fomin, V.G.; Lopez, Y.S.; et al. Genetic Variation in Choline-Metabolizing Enzymes Alters Choline Metabolism in Young Women Consuming Choline Intakes Meeting Current Recommendations. Int. J. Mol. Sci. 2017, 18, 252. https://doi.org/10.3390/ijms18020252
Ganz AB, Cohen VV, Swersky CC, Stover J, Vitiello GA, Lovesky J, Chuang JC, Shields K, Fomin VG, Lopez YS, et al. Genetic Variation in Choline-Metabolizing Enzymes Alters Choline Metabolism in Young Women Consuming Choline Intakes Meeting Current Recommendations. International Journal of Molecular Sciences. 2017; 18(2):252. https://doi.org/10.3390/ijms18020252
Chicago/Turabian StyleGanz, Ariel B., Vanessa V. Cohen, Camille C. Swersky, Julie Stover, Gerardo A. Vitiello, Jessica Lovesky, Jasmine C. Chuang, Kelsey Shields, Vladislav G. Fomin, Yusnier S. Lopez, and et al. 2017. "Genetic Variation in Choline-Metabolizing Enzymes Alters Choline Metabolism in Young Women Consuming Choline Intakes Meeting Current Recommendations" International Journal of Molecular Sciences 18, no. 2: 252. https://doi.org/10.3390/ijms18020252





