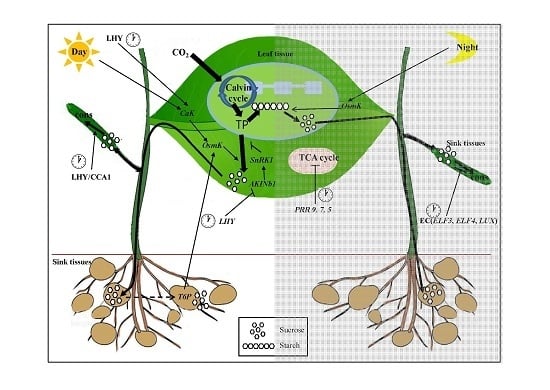
The Importance of the Circadian Clock in Regulating Plant Metabolism
Abstract
:1. Introduction
2. Circadian Oscillation of Primary (Carbon) Metabolites
3. Regulation of Starch Metabolites by the Circadian Clock
4. Regulation of Circadian-Mediated Carbon Productivity in Crops
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [PubMed]
- Farre, E.M.; Weise, S.E. The interactions between the circadian clock and primary metabolism. Curr. Opin. Plant Biol. 2012, 15, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Brody, S. The Genetics of Circadian Rhythms, 1st ed.; Academic Press: San Diego, CA, USA, 2011; p. 254. [Google Scholar]
- Francis, D. Observations on stomata. Philos. Trans. R. Soc. Lond. B 1898, 190, 531–621. [Google Scholar]
- McClung, C.R. Plant circadian rhythms. Plant Cell 2006, 18, 792–803. [Google Scholar] [CrossRef] [PubMed]
- Covington, M.F.; Panda, S.; Liu, X.L.; Strayer, C.A.; Wagner, D.R.; Kay, S.A. ELF3 modulates resetting of the circadian clock in Arabidopsis. Plant Cell 2001, 13, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- McClung, C.R. Comes a time. Curr. Opin. Plant Biol. 2008, 11, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Andersson, C.R.; Kondo, T.; Golden, S.S.; Johnson, C.H. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA 1998, 95, 8660–8664. [Google Scholar] [CrossRef] [PubMed]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kevei, E.; Toth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef] [PubMed]
- Espinoza, C.; Degenkolbe, T.; Caldana, C.; Zuther, E.; Leisse, A.; Willmitzer, L.; Hincha, D.K.; Hannah, M.A. Interaction with Diurnal and Circadian Regulation Results in Dynamic Metabolic and Transcriptional Changes during Cold Acclimation in Arabidopsis. PLoS ONE 2010, 5, e14101. [Google Scholar] [CrossRef] [PubMed]
- Greenham, K.; McClung, C.R. Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 2015, 16, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Ebenhoh, O. Mathematical modelling of diurnal regulation of carbohydrate allocation by osmo-related processes in plants. J. R. Soc. Interface 2015, 12, 20141357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Smith, A.M. Starch and the clock: The dark side of plant productivity. Trends Plant Sci. 2011, 16, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Pantin, F.; Simonneau, T.; Rolland, G.; Dauzat, M.; Muller, B. Control of Leaf Expansion: A Developmental Switch from Metabolics to Hydraulics. Plant Physiol. 2011, 156, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Gibon, Y.; Lunn, J.E.; Piques, M. Multilevel genomics analysis of carbon signalling during low carbon availability: Coordinating the supply and utilisation of carbon in a fluctuating environment. Funct. Plant Biol. 2007, 34, 526–549. [Google Scholar] [CrossRef]
- Kotting, O.; Kossmann, J.; Zeeman, S.C.; Lloyd, J.R. Regulation of starch metabolism: The age of enlightenment? Curr. Opin. Plant Biol. 2010, 13, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Gehan, J.P.; Sharkey, T.D. Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 2005, 138, 2280–2291. [Google Scholar] [CrossRef] [PubMed]
- Tenorio, G.; Orea, A.; Romero, J.M.; Merida, A. Oscillation of mRNA level and activity of granule-bound starch synthase I in Arabidopsis leaves during the day/night cycle. Plant Mol. Biol. 2003, 51, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.J.; Yeh, K.W.; Tsai, C.Y. Regulation of starch granule-bound starch synthase I gene expression by circadian clock and sucrose in the source tissue of sweet potato. Plant Sci. 2001, 161, 635–644. [Google Scholar] [CrossRef]
- Rikin, A.; Dillwith, J.W.; Bergman, D.K. Correlation between the Circadian-Rhythm of Resistance to Extreme Temperatures and Changes in Fatty-Acid Composition in Cotton Seedlings. Plant Physiol. 1993, 101, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Van Harsselaar, J.K.; Lorenz, J.; Senning, M.; Sonnewald, U.; Sonnewald, S. Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genom. 2017, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Geigenberger, P.; Stitt, M. Diurnal changes in sucrose, nucleotides, starch synthesis and AGPS transcript in growing potato tubers that are suppressed by decreased expression of sucrose phosphate synthase. Plant J. 2000, 23, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Haydon, M.J.; Hearn, T.J.; Bell, L.J.; Hannah, M.A.; Webb, A.A. Metabolic regulation of circadian clocks. Semin. Cell Dev. Biol. 2013, 24, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Geiger, D.R.; Shieh, W.J. Evidence for circadian regulation of starch and sucrose synthesis in sugar beet leaves. Plant Physiol. 1992, 99, 1393–1399. [Google Scholar] [CrossRef] [PubMed]
- Merida, A.; Rodriguez-Galan, J.M.; Vincent, C.; Romero, J.M. Expression of the granule-bound starch synthase I (Waxy) gene from snapdragon is developmentally and circadian clock regulated. Plant Physiol. 1999, 120, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Halford, N.G.; Hey, S.; Jhurreea, D.; Laurie, S.; McKibbin, R.S.; Paul, M.; Zhang, Y. Metabolic signalling and carbon partitioning: Role of Snf1-related (SnRK1) protein kinase. J. Exp. Bot. 2003, 54, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Sugden, C.; Donaghy, P.G.; Halford, N.G.; Hardie, D.G. Two SNF1-Related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999, 120, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Kulma, A.; Villadsen, D.; Campbell, D.G.; Meek, S.E.M.; Harthill, J.E.; Nielsen, T.H.; MacKintosh, C. Phosphorylation and 14-3-3 binding of Arabidopsis 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase. Plant J. 2004, 37, 654–667. [Google Scholar] [CrossRef] [PubMed]
- Robertson, F.; Skeffington, A.; Gardner, M.; Webb, A.A.R. Interactions between circadian and hormonal signalling in plants. Plant Mol. Biol. 2009, 69, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Pokhilko, A.; Mas, P.; Millar, A.J. Modelling the widespread effects of TOC1 signalling on the plant circadian clock and its outputs. BMC Syst. Biol. 2013, 7, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pokhilko, A.; Flis, A.; Sulpice, R.; Stitt, M.; Ebenhoh, O. Adjustment of carbon fluxes to light conditions regulates the daily turnover of starch in plants: A computational model. Mol. Biosyst. 2014, 10, 613–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niwa, Y.; Yamashino, T.; Mizuno, T. The Circadian Clock Regulates the Photoperiodic Response of Hypocotyl Elongation through a Coincidence Mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 838–854. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.T.; Koo, B.H.; Kim, D.; Yoo, S.C.; Paek, N.C. Casein Kinases I and 2 alpha Phosphorylate Oryza Sativa Pseudo-Response Regulator 37 (OsPRR37) in Photoperiodic Flowering in Rice. Mol. Cells 2015, 38, 81–88. [Google Scholar] [PubMed]
- Mizoguchi, T.; Wheatley, K.; Hanzawa, Y.; Wright, L.; Mizoguchi, M.; Song, H.R.; Carre, I.A.; Coupland, G. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2002, 2, 629–641. [Google Scholar] [CrossRef]
- Alabadi, D.; Oyama, T.; Yanovsky, M.J.; Harmon, F.G.; Mas, P.; Kay, S.A. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 2001, 293, 880–883. [Google Scholar] [CrossRef] [PubMed]
- Chew, Y.H.; Wenden, B.; Flis, A.; Mengin, V.; Taylor, J.; Davey, C.L.; Tindal, C.; Thomas, H.; Ougham, H.J.; de Reffye, P.; et al. Multiscale digital Arabidopsis predicts individual organ and whole-organism growth. Proc. Natl. Acad. Sci. USA 2014, 111, E4127–E4136. [Google Scholar] [CrossRef] [PubMed]
- Yazdanbakhsh, N.; Sulpice, R.; Graf, A.; Stitt, M.; Fisahn, J. Circadian control of root elongation and C partitioning in Arabidopsis thaliana. Plant Cell Environ. 2011, 34, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognar, L.; Nagy, F.; Millar, A.J.; Amasino, R.M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Dowson-Day, M.J.; Millar, A.J. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J. 1999, 17, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Nozue, K.; Covington, M.F.; Duek, P.D.; Lorrain, S.; Fankhauser, C.; Harmer, S.L.; Maloof, J.N. Rhythmic growth explained by coincidence between internal and external cues. Nature 2007, 448, 358–361. [Google Scholar] [CrossRef] [PubMed]
- Kehr, J.; Hustiak, F.; Walz, C.; Willmitzer, L.; Fisahn, J. Transgenic plants changed in carbon allocation pattern display a shift in diurnal growth pattern. Plant J. 1998, 16, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Hotta, C.T.; Gardner, M.J.; Hubbard, K.E.; Baek, S.J.; Dalchau, N.; Suhita, D.; Dodd, A.N.; Webb, A.A. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007, 30, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, A.; Kusano, M.; Nakamichi, N.; Kobayashi, M.; Hayashi, N.; Sakakibara, H.; Mizuno, T.; Saito, K. Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc. Natl. Acad. Sci. USA 2009, 106, 7251–7256. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.D.; Morris, W.L.; Ducreux, L.J.; Morris, J.A.; Usman, M.; Verrall, S.R.; Fuller, J.; Simpson, C.G.; Zhang, R.; Hedley, P.E.; et al. Physiological, biochemical and molecular responses of the potato (Solanum tuberosum L.) plant to moderately elevated temperature. Plant Cell Environ. 2014, 37, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Poorter, H.; Nagel, O. The role of biomass allocation in the growth response of plants to different levels of light, CO2, nutrients and water: A quantitative review. Aust. J. Plant Physiol. 2000, 27, 1191. [Google Scholar] [CrossRef]
- Zeeman, S.C.; Kossmann, J.; Smith, A.M. Starch: Its Metabolism, Evolution, and Biotechnological Modification in Plants. Ann. Rev. Plant Biol. 2010, 61, 209–234. [Google Scholar] [CrossRef] [PubMed]
- Tsai, H.L.; Lue, W.L.; Lu, K.J.; Hsieh, M.H.; Wang, S.M.; Chen, J. Starch Synthesis in Arabidopsis Is Achieved by Spatial Cotranscription of Core Starch Metabolism Genes. Plant Physiol. 2009, 151, 1582–1595. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.R.; McHale, N.A. A Starchless Mutant of Nicotiana sylvestris Containing a Modified Plastid Phosphoglucomutase. Plant Physiol. 1988, 88, 838–844. [Google Scholar] [CrossRef] [PubMed]
- Gagic, M.; Faville, M.; Kardailsky, I.; Putterill, J. Comparative Genomics and Functional Characterisation of the GIGANTEA Gene from the Temperate Forage Perennial Ryegrass Lolium perenne. Plant Mol. Biol. Rep. 2015, 33, 1098–1106. [Google Scholar] [CrossRef]
- Kim, J.A.; Jung, H.E.; Hong, J.K.; Hermand, V.; McClung, C.R.; Lee, Y.H.; Kim, J.Y.; Lee, S.I.; Jeong, M.J.; Kim, J.; et al. Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 2016, 35, 1943–1954. [Google Scholar] [CrossRef] [PubMed]
- Thiruvengadam, M.; Shih, C.F.; Yang, C.H. Expression of An Antisense Brassica oleracea GIGANTEA (BoGI) Gene in Transgenic Broccoli Causes Delayed Flowering, Leaf Senescence, and Post-Harvest Yellowing Retardation. Plant Mol. Biol. Rep. 2015, 33, 1499–1509. [Google Scholar] [CrossRef]
- Tang, W.; Yan, H.; Su, Z.X.; Park, S.C.; Liu, Y.J.; Zhang, Y.G.; Wang, X.; Kou, M.; Ma, D.F.; Kwak, S.S.; et al. Cloning and characterization of a novel GIGANTEA gene in sweet potato. Plant Physiol. Biochem. 2017, 116, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Ashikari, M.; Miura, K.; Yamashino, T.; Mizuno, T. The evolutionarily conserved OsPRR quintet: Rice pseudo-response regulators implicated in circadian rhythm. Plant Cell Physiol. 2003, 44, 1229–1236. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Mihara, M.; Suzuki, Y.; Gupta, M.; Itoh, H.; Nagano, A.J.; Motoyama, R.; Sawada, Y.; Yano, M.; Hirai, M.Y.; et al. Os-GIGANTEA confers robust diurnal rhythms on the global transcriptome of rice in the field. Plant Cell 2011, 23, 1741–1755. [Google Scholar] [CrossRef] [PubMed]
- Abbo, S.; Van-Oss, R.P.; Gopher, A.; Saranga, Y.; Ofner, R.; Peleg, Z. Plant domestication versus crop evolution: A conceptual framework for cereals and grain legumes. Trends Plant Sci. 2014, 19, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef] [PubMed]
- DeVries, I.M. Origin and domestication of Lactuca sativa L. Genet. Res. Crop. Evol. 1997, 44, 165–174. [Google Scholar] [CrossRef]
- Yarkhunova, Y.; Edwards, C.E.; Ewers, B.E.; Baker, R.L.; Aston, T.L.; McClung, C.R.; Lou, P.; Weinig, C. Selection during crop diversification involves correlated evolution of the circadian clock and ecophysiological traits in Brassica rapa. New Phytol. 2016, 210, 133–144. [Google Scholar] [CrossRef] [PubMed]
- Navarro, C.; Abelenda, J.A.; Cruz-Oro, E.; Cuellar, C.A.; Tamaki, S.; Silva, J.; Shimamoto, K.; Prat, S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 2011, 478, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Kim, J.S.; Hong, J.K.; Lee, Y.H.; Choi, B.S.; Seol, Y.J.; Jeon, C.H. Comparative mapping, genomic structure, and expression analysis of eight pseudo-response regulator genes in Brassica rapa. Mol. Genet. Gen. 2012, 287, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Yang, T.J.; Kim, J.S.; Park, J.Y.; Kwon, S.J.; Lim, M.H.; Jin, M.; Lee, S.C.; Lee, S.I.; Choi, B.S.; et al. Isolation of circadian-associated genes in Brassica rapa by comparative genomics with Arabidopsis thaliana. Mol. Cells 2007, 23, 145–153. [Google Scholar] [PubMed]
- Coello, P.; Hirano, E.; Hey, S.J.; Muttucumaru, N.; Martinez-Barajas, E.; Parry, M.A.J.; Halford, N.G. Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK) 1 in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 2012, 63, 913–924. [Google Scholar] [CrossRef] [PubMed]
- Yakir, E.; Hilman, D.; Harir, Y.; Green, R.M. Regulation of output from the plant circadian clock. FEBS J. 2007, 274, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.; Bledsoe, S.W.; Siekman, A.; Kollman, A.; Waters, B.M.; Feil, R.; Stitt, M.; Lagrimini, L.M. The trehalose pathway in maize: Conservation and gene regulation in response to the diurnal cycle and extended darkness. J. Exp. Bot. 2014, 65, 5959–5973. [Google Scholar] [CrossRef] [PubMed]
- Ruuska, S.A.; Schwender, J.; Ohlrogge, J.B. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 2004, 136, 2700–2709. [Google Scholar] [CrossRef] [PubMed]
- Goffman, F.D.; Alonso, A.P.; Schwender, J.; Shachar-Hill, Y.; Ohlrogge, J.B. Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 2005, 138, 2269–2279. [Google Scholar] [CrossRef] [PubMed]
- Faure, S.; Turner, A.S.; Gruszka, D.; Christodoulou, V.; Davis, S.J.; von Korff, M.; Laurie, D.A. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc. Natl. Acad. Sci. USA 2012, 109, 8328–8333. [Google Scholar] [CrossRef] [PubMed]
- Dalchau, N.; Baek, S.J.; Briggs, H.M.; Robertson, F.C.; Dodd, A.N.; Gardner, M.J.; Stancombe, M.A.; Haydon, M.J.; Stan, G.B.; Goncalves, J.M.; et al. The circadian oscillator gene GIGANTEA mediates a long-term response of the Arabidopsis thaliana circadian clock to sucrose. Proc. Natl. Acad. Sci. USA 2011, 108, 5104–5109. [Google Scholar] [CrossRef] [PubMed]
- Bolouri Moghaddam, M.R.; Van den Ende, W. Sweet immunity in the plant circadian regulatory network. J. Exp. Bot. 2013, 64, 1439–1449. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Ye, M.; Jiang, S. Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 2005, 24, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kusano, M.; Fukushima, A.; Kita, M.; Ito, S.; Yamashino, T.; Saito, K.; Sakakibara, H.; Mizuno, T. Transcript profiling of an Arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009, 50, 447–462. [Google Scholar] [CrossRef] [PubMed]
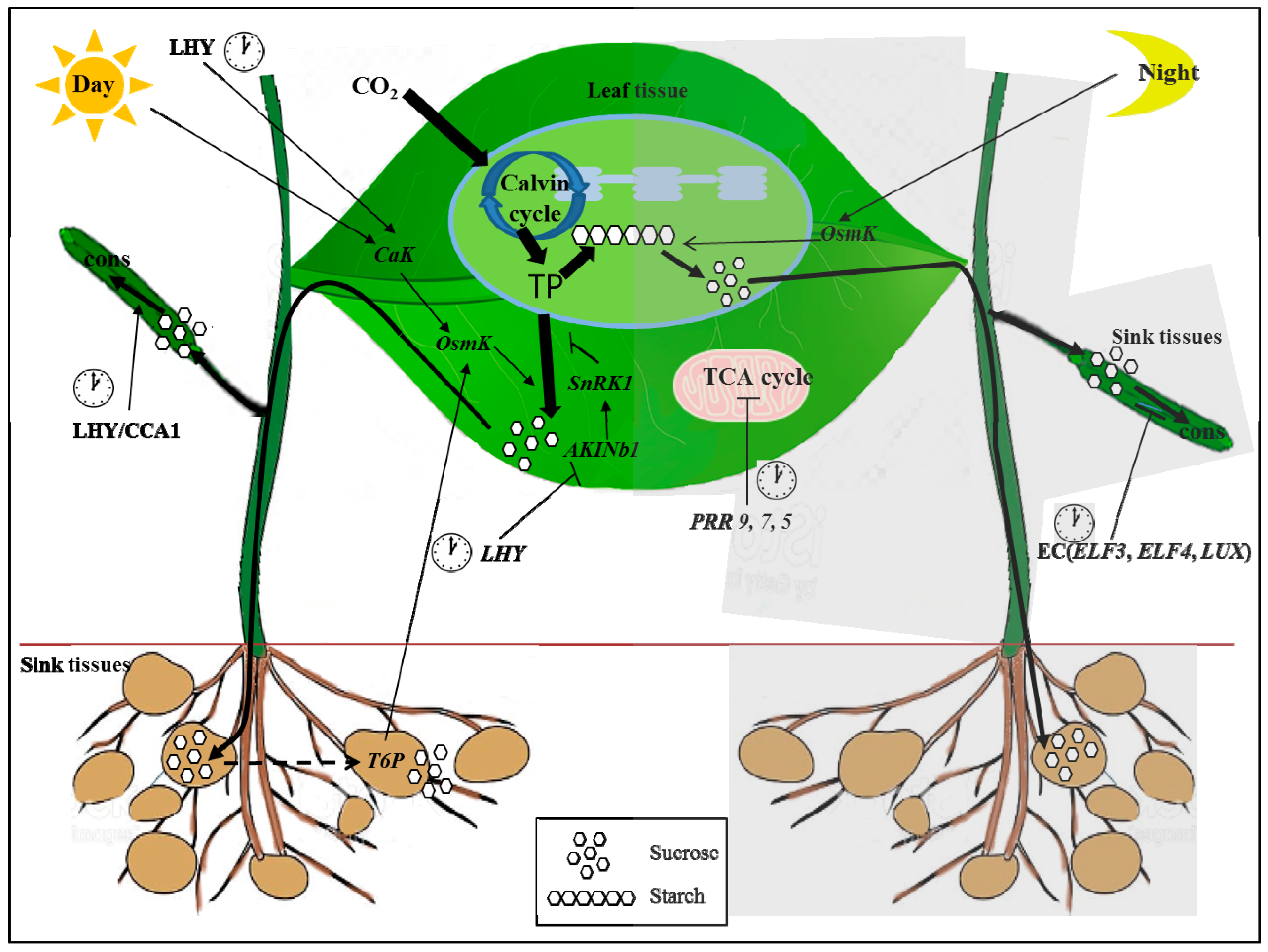
| Name of Gene | Arabidopsis | Homologus Genes in the Crops | Reference |
|---|---|---|---|
| Clock genes | |||
| Late elongated hypocotyl (LHY) | AT1G01060 | OS-LHY (Oryza sativa, rice) LHY (Solanum tuberosum, potato) | Izawa et al., 2011 [57] Hancock et al., 2014 [44] |
| Circadian clock associated 1 (CCA1) | AT2G46830 | N.A.* | |
| Early Flowering 3 (ELF3) | AT2G25930 | OsELF3 (Oryza sativa, rice) | Kwon et al., 2015 [35] |
| Early Flowering 4 (ELF4) | AT2G40080 | OsELF4 (Oryza sativa, rice) | |
| Lux arrhythmo (LUX) | AT3G46640 | OsLUX (Oryza sativa, rice) | Kwon et al., 2015 [35] |
| Pseudo-response regulator (PRR)s | AT1G32100 AT5G60100 AT5G24470 AT5G02810 AT2G46790 | OsPRRs (Oryza sativa, rice) BrPRRs (Brassica rapa, Chinese cabbage) | Murakami et al., 2003 [56] Kim et al., 2007, 2012 [63,64] |
| GIGANTEA (GI) | AT1G22770 | OsGI (Oryza sativa, rice) StGIa,b (Solanum tuberosum, potato) GmGIa (Glycine max, soybean) IbGI (Ipomoea batatas Lam, sweet potato) BoGI (Barssica oleracea L. broccoli) BrGI (Brassica rapa, Chinese cabbage) LpGI (Lolium perenne, ryegrass) | Kwon et al., 2015 [35] Hancock et al., 2014 [44] Watanabe et al., 2011 [55] Tang et al., 2017 [54] Thiruvengadam et al., 2015 [53] Kim et al., 2016 [52] Gagic et al., 2015 [51] |
| Clock sensing genes | |||
| Ca2+-dependent kinase (CaK) | N.A. | ||
| SNF1-related protein kinase 1 (SnRK1) | AT5G39440 | SnRK1 (Spinacia oleracea, spinach) SnRK1 (Nicotiana tabacum, tobacco) SnRK1 (Secale cereale, rye) SnRK1 (Solanum tuberosum, potato) SnRK1 (Hordeum vulgare barley) SnRK1 (Triticum aestivum, wheat) | Sugden et al., 1999 [29] Sugden et al., 1999 [29] Sugden et al., 1999 [29] Sugden et al., 1999 [29] Halford et al., 2003 [28] Coello et al., 2012 [65] |
| ARABIDOPSIS KINASE (AKIN) b1 | AT5G21170 | N.A. | |
| osmo-sensitive kinase (OsmK) | N.A | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.A.; Kim, H.-S.; Choi, S.-H.; Jang, J.-Y.; Jeong, M.-J.; Lee, S.I. The Importance of the Circadian Clock in Regulating Plant Metabolism. Int. J. Mol. Sci. 2017, 18, 2680. https://doi.org/10.3390/ijms18122680
Kim JA, Kim H-S, Choi S-H, Jang J-Y, Jeong M-J, Lee SI. The Importance of the Circadian Clock in Regulating Plant Metabolism. International Journal of Molecular Sciences. 2017; 18(12):2680. https://doi.org/10.3390/ijms18122680
Chicago/Turabian StyleKim, Jin A, Hyun-Soon Kim, Seo-Hwa Choi, Ji-Young Jang, Mi-Jeong Jeong, and Soo In Lee. 2017. "The Importance of the Circadian Clock in Regulating Plant Metabolism" International Journal of Molecular Sciences 18, no. 12: 2680. https://doi.org/10.3390/ijms18122680