CMG2 Expression Is an Independent Prognostic Factor for Soft Tissue Sarcoma Patients
Abstract
:1. Introduction
2. Results
2.1. CMG2 mRNA and Protein Expression in Soft Tissue Sarcoma
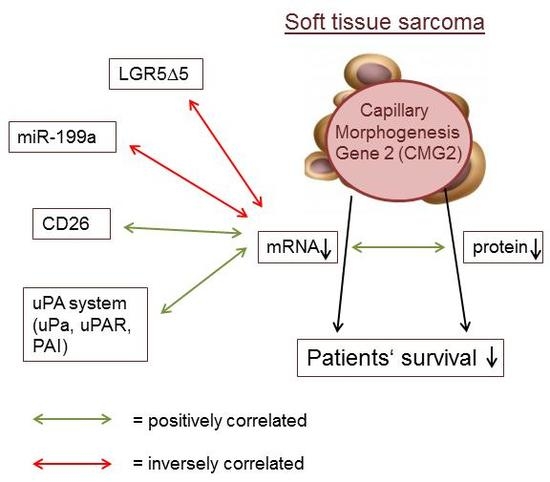
2.2. Association of CMG2 mRNA to Angiogenesis- and Metastasis-Related Genes
2.3. CMG2 Expression and Patient Survival
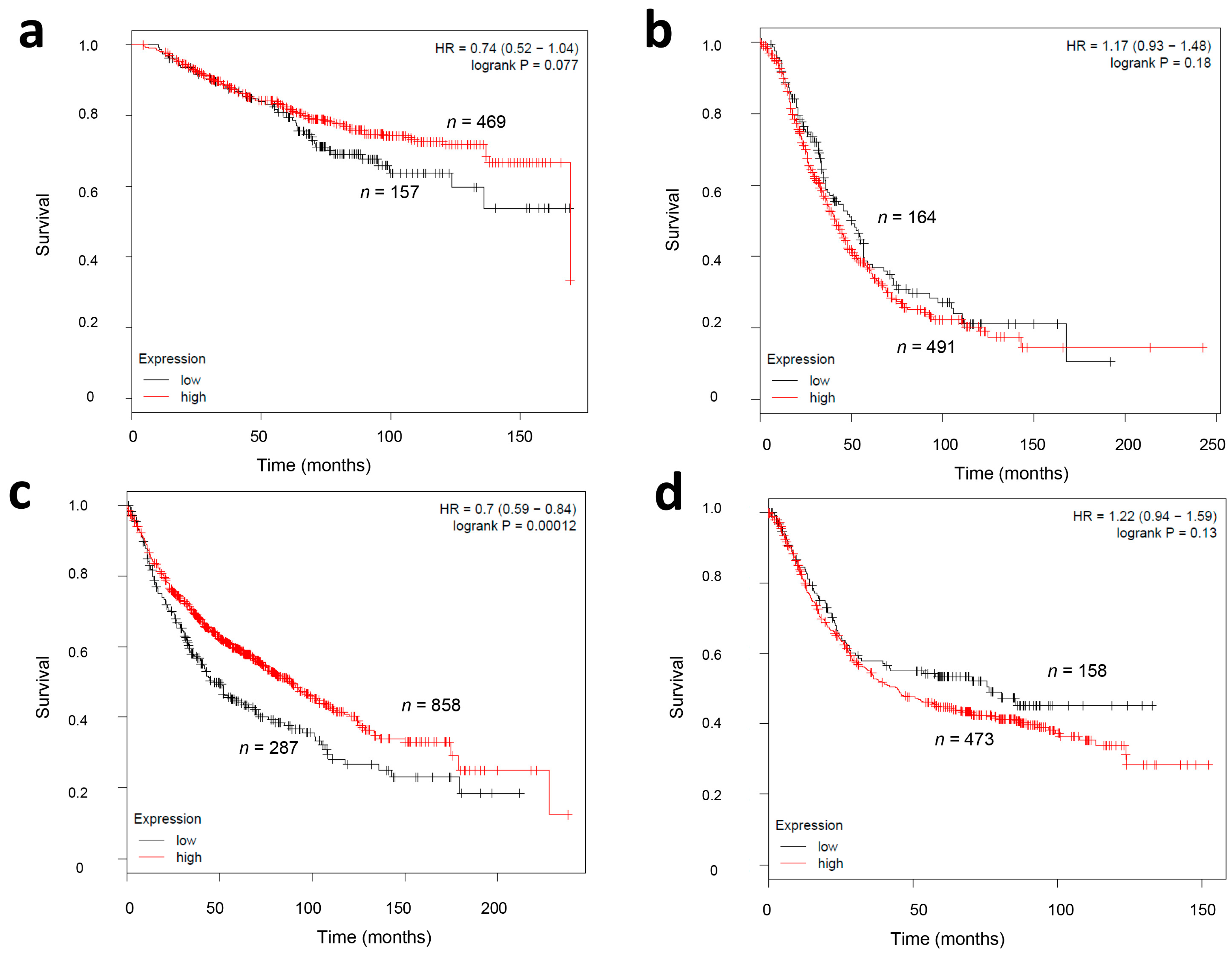
2.4. CMG2 mRNA Expression and Survival in Other Tumor Entities
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. RNA Isolation
4.3. cDNA Synthesis and qPCR
4.4. Protein Isolation and ELISA
4.5. Statistics
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Katenkamp, D.; Kosmehl, H. Heterogeneity in malignant soft tissue tumors. Curr. Top. Pathol. 1995, 89, 123–151. [Google Scholar] [PubMed]
- Wu, C.; Wei, Q.; Utomo, V.; Nadesan, P.; Whetstone, H.; Kandel, R.; Wunder, J.S.; Alman, B.A. Side population cells isolated from mesenchymal neoplasms have tumor initiating potential. Cancer Res. 2007, 67, 8216–8222. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.A. Sarcoma classification: An update based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone. Cancer 2014, 120, 1763–1774. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, A.; Sultan, I.; Huang, T.T.; Rodriguez-Galindo, C.; Shehadeh, A.; Meazza, C.; Ness, K.K.; Casanova, M.; Spunt, S.L. Soft tissue sarcoma across the age spectrum: A population-based study from the Surveillance Epidemiology and End Results database. Pediatr. Blood Cancer 2011, 57, 943–949. [Google Scholar] [CrossRef] [PubMed]
- Steinestel, K.; Wardelmann, E. Metastasierung und Progressionsmechanismen von Weichteiltumoren. Der Pathologe 2015, 36, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Pisters, P.W.; Leung, D.H.; Woodruff, J.; Shi, W.; Brennan, M.F. Analysis of prognostic factors in 1041 patients with localized soft tissue sarcomas of the extremities. J. Clin. Oncol. 1996, 14, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.E.; Mavila, A.; Salazar, R.; Bayless, K.J.; Kanagala, S.; Maxwell, S.A.; Davis, G.E. Differential gene expression during capillary morphogenesis in 3D collagen matrices: Regulated expression of genes involved in basement membrane matrix assembly, cell cycle progression, cellular differentiation and G-protein signaling. J. Cell Sci. 2001, 114, 2755–2773. [Google Scholar] [PubMed]
- Scobie, H.M.; Rainey, G.J.A.; Bradley, K.A.; Young, J.A.T. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc. Natl. Acad. Sci. USA 2003, 100, 5170–5174. [Google Scholar] [CrossRef] [PubMed]
- Hanks, S.; Adams, S.; Douglas, J.; Arbour, L.; Atherton, D.J.; Balci, S.; Bode, H.; Campbell, M.E.; Feingold, M.; Keser, G.; et al. Mutations in the gene encoding capillary morphogenesis protein 2 cause juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am. J. Hum. Genet. 2003, 73, 791–800. [Google Scholar] [CrossRef] [PubMed]
- Dowling, O.; Difeo, A.; Ramirez, M.C.; Tukel, T.; Narla, G.; Bonafe, L.; Kayserili, H.; Yuksel-Apak, M.; Paller, A.S.; Norton, K.; et al. Mutations in capillary morphogenesis gene-2 result in the allelic disorders juvenile hyaline fibromatosis and infantile systemic hyalinosis. Am. J. Hum. Genet. 2003, 73, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Deuquet, J.; Abrami, L.; Difeo, A.; Ramirez, M.C.M.; Martignetti, J.A.; van der Goot, F.G. Systemic hyalinosis mutations in the CMG2 ectodomain leading to loss of function through retention in the endoplasmic reticulum. Hum. Mutat. 2009, 30, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Deuquet, J.; Lausch, E.; Superti-Furga, A.; van der Goot, F.G. The dark sides of capillary morphogenesis gene 2. EMBO J. 2012, 31, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Bürgi, J.; Kunz, B.; Abrami, L.; Deuquet, J.; Piersigilli, A.; Scholl-Bürgi, S.; Lausch, E.; Unger, S.; Superti-Furga, A.; Bonaldo, P.; et al. CMG2/ANTXR2 regulates extracellular collagen VI which accumulates in hyaline fibromatosis syndrome. Nat. Commun. 2017, 8, 15861. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Sun, P.-H.; Malik, M.F.A.; Mason, M.D.; Jiang, W.G. Capillary morphogenesis gene 2 inhibits growth of breast cancer cells and is inversely correlated with the disease progression and prognosis. J. Cancer Res. Clin. Oncol. 2014, 140, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Sanders, A.J.; Sun, P.-H.; Mason, M.D.; Jiang, W.G. Capillary morphogenesis gene 2 regulates adhesion and invasiveness of prostate cancer cells. Oncol. Lett. 2014, 7, 2149–2153. [Google Scholar] [CrossRef] [PubMed]
- Dass, K.; Ahmad, A.; Azmi, A.S.; Sarkar, S.H.; Sarkar, F.H. Evolving role of uPA/uPAR system in human cancers. Cancer Treat. Rev. 2008, 34, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Noh, H.; Hong, S.; Huang, S. Role of urokinase receptor in tumor progression and development. Theranostics 2013, 3, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Seetoo, D.Q.; Crowe, P.J.; Russell, P.J.; Yang, J.L. Quantitative expression of protein markers of plasminogen activation system in prognosis of colorectal cancer. J. Surg. Oncol. 2003, 82, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Scarlett, C.J.; Jackson, C.J.; Allen, B.J.; Smith, R.C. Prognostic significance of growth factors and the urokinase-type plasminogen activator system in pancreatic ductal adenocarcinoma. Pancreas 2008, 36, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Foekens, J.A.; Peters, H.A.; Look, M.P.; Portengen, H.; Schmitt, M.; Kramer, M.D.; Brünner, N.; Jänicke, F.; Meijer-van Gelder, M.E.; Henzen-Logmans, S.C.; et al. The urokinase system of plasminogen activation and prognosis in 2780 breast cancer patients. Cancer Res. 2000, 60, 636–643. [Google Scholar] [PubMed]
- Taubert, H.; Magdolen, V.; Kotzsch, M. Impact of expression of the uPA system in sarcomas. Biomark. Med. 2013, 7, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Kotzsch, M.; Magdolen, V.; Greither, T.; Kappler, M.; Bache, M.; Lautenschläger, C.; Füssel, S.; Eckert, A.W.; Luther, T.; Baretton, G.; et al. Combined mRNA expression levels of members of the urokinase plasminogen activator (uPA) system correlate with disease-associated survival of soft-tissue sarcoma patients. BMC Cancer 2011, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Rot, S.; Taubert, H.; Bache, M.; Greither, T.; Würl, P.; Eckert, A.W.; Schubert, J.; Vordermark, D.; Kappler, M. A novel splice variant of the stem cell marker LGR5/GPR49 is correlated with the risk of tumor-related death in soft-tissue sarcoma patients. BMC Cancer 2011, 11, 429. [Google Scholar] [CrossRef] [PubMed]
- Keßler, J.; Rot, S.; Bache, M.; Kappler, M.; Würl, P.; Vordermark, D.; Taubert, H.; Greither, T. miR-199a-5p regulates HIF-1α and OSGIN2 and its expression is correlated to soft-tissue sarcoma patients’ outcome. Oncol. Lett. 2016, 12, 5281–5288. [Google Scholar] [PubMed]
- Lessard, J.; Pelletier, M.; Biertho, L.; Biron, S.; Marceau, S.; Hould, F.-S.; Lebel, S.; Moustarah, F.; Lescelleur, O.; Marceau, P.; et al. Characterization of dedifferentiating human mature adipocytes from the visceral and subcutaneous fat compartments: Fibroblast-activation protein alpha and dipeptidyl peptidase 4 as major components of matrix remodeling. PLoS ONE 2015, 10, e0122065. [Google Scholar] [CrossRef] [PubMed]
- Dohi, O.; Ohtani, H.; Hatori, M.; Sato, E.; Hosaka, M.; Nagura, H.; Itoi, E.; Kokubun, S. Histogenesis-specific expression of fibroblast activation protein and dipeptidylpeptidase-IV in human bone and soft tissue tumours. Histopathology 2009, 55, 432–440. [Google Scholar] [CrossRef] [PubMed]
- Benassi, M.S.; Ponticelli, F.; Azzoni, E.; Gamberi, G.; Pazzaglia, L.; Chiechi, A.; Conti, A.; Spessotto, P.; Scapolan, M.; Pignotti, E.; et al. Altered expression of urokinase-type plasminogen activator and plasminogen activator inhibitor in high-risk soft tissue sarcomas. Histol. Histopathol. 2007, 22, 1017–1024. [Google Scholar] [PubMed]
- Taubert, H.; Würl, P.; Greither, T.; Kappler, M.; Bache, M.; Lautenschläger, C.; Füssel, S.; Meye, A.; Eckert, A.W.; Holzhausen, H.-J.; et al. Co-detection of members of the urokinase plasminogen activator system in tumour tissue and serum correlates with a poor prognosis for soft-tissue sarcoma patients. Br. J. Cancer 2010, 102, 731–737. [Google Scholar] [CrossRef] [PubMed]
- Garcia, M.I.; Ghiani, M.; Lefort, A.; Libert, F.; Strollo, S.; Vassart, G. LGR5 deficiency deregulates Wnt signaling and leads to precocious Paneth cell differentiation in the fetal intestine. Dev. Biol. 2009, 331, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Tanese, K.; Fukuma, M.; Yamada, T.; Mori, T.; Yoshikawa, T.; Watanabe, W.; Ishiko, A.; Amagai, M.; Nishikawa, T.; Sakamoto, M. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am. J. Pathol. 2008, 173, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Uchida, H.; Yamazaki, K.; Fukuma, M.; Yamada, T.; Hayashida, T.; Hasegawa, H.; Kitajima, M.; Kitagawa, Y.; Sakamoto, M. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci. 2010, 101, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Merlos-Suárez, A.; Barriga, F.M.; Jung, P.; Iglesias, M.; Céspedes, M.V.; Rossell, D.; Sevillano, M.; Hernando-Momblona, X.; da Silva-Diz, V.; Muñoz, P.; et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell 2011, 8, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Ryuge, S.; Sato, Y.; Jiang, S.-X.; Wang, G.; Kobayashi, M.; Nagashio, R.; Katono, K.; Iyoda, A.; Satoh, Y.; Masuda, N. The clinicopathological significance of Lgr5 expression in lung adenocarcinoma. Lung Cancer 2013, 82, 143–148. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, H.; Zhang, J.; Zhai, L.; Chen, W.; Zhao, C. miR-199a-5p regulates β1 integrin through Ets-1 to suppress invasion in breast cancer. Cancer Sci. 2016, 107, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fan, K.-J.; Sun, Q.; Chen, A.-Z.; Shen, W.-L.; Zhao, Z.-H.; Zheng, X.-F.; Yang, X. Functional screening for miRNAs targeting Smad4 identified miR-199a as a negative regulator of TGF-β signalling pathway. Nucleic Acids Res. 2012, 40, 9286–9297. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.-G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, R.; Wang, Y.; Tang, J.; Tang, S.; Chen, X.; Xia, K.; Xiong, W.; Xu, D.; Wang, S.; et al. miR-199a-5p regulates the expression of metastasis-associated genes in B16F10 melanoma cells. Int. J. Clin. Exp. Pathol. 2014, 7, 7182–7190. [Google Scholar] [PubMed]
- Si, T.; Liu, C.; Xu, K.; Gui, Y. Association of miR-199a expression with clinicopathologic characteristics and prognosis of renal cell carcinoma. J. South. Med. Univ. 2012, 32, 1568–1571. [Google Scholar]
- Gu, S.; Cheung, H.H.; Lee, T.L.; Lu, G.; Poon, W.S.; Chan, W.Y. Molecular mechanisms of regulation and action of microRNA-199a in testicular germ cell tumor and glioblastomas. PLoS ONE 2013, 8, e83980. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Duan, H.; Zhou, S.; Liu, Z.; Wu, D.; Zhao, T.; Xu, S.; Yang, L.; Li, D. microRNA-199a-3p functions as tumor suppressor by regulating glucose metabolism in testicular germ cell tumors. Mol. Med. Rep. 2016, 14, 2311–2320. [Google Scholar] [CrossRef] [PubMed]
- Salamone, M.; Carfì Pavia, F.; Ghersi, G. Proteolytic Enzymes Clustered in Specialized Plasma-Membrane Domains Drive Endothelial Cells’ Migration. PLoS ONE 2016, 11, e0154709. [Google Scholar] [CrossRef] [PubMed]
- Reeves, C.V.; Dufraine, J.; Young, J.A.T.; Kitajewski, J. Anthrax toxin receptor 2 is expressed in murine and tumor vasculature and functions in endothelial proliferation and morphogenesis. Oncogene 2010, 29, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.E.; Zhang, Y.; Molinolo, A.A.; Miller-Randolph, S.; Szabo, R.; Bugge, T.H.; Leppla, S.H.; Liu, S. Capillary morphogenesis protein-2 is required for mouse parturition by maintaining uterine collagen homeostasis. Biochem. Biophys. Res. Commun. 2012, 422, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Cryan, L.; Habeshian, K.A.; Murillo, C.; Tamayo-Castillo, G.; Rogers, M.S.; Clardy, J. Phenolic compounds as antiangiogenic CMG2 inhibitors from Costa Rican endophytic fungi. Bioorg. Med. Chem. Lett. 2012, 22, 5885–5888. [Google Scholar] [CrossRef] [PubMed]
- Cryan, L.M.; Bazinet, L.; Habeshian, K.A.; Cao, S.; Clardy, J.; Christensen, K.A.; Rogers, M.S. 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranose inhibits angiogenesis via inhibition of capillary morphogenesis gene 2. J. Med. Chem. 2013, 56, 1940–1945. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Sun, P.-H.; Sanders, A.J.; Martin, T.A.; Lane, J.; Mason, M.D.; Jiang, W.G. Therapeutic potential of capillary morphogenesis gene 2 extracellular vWA domain in tumour-related angiogenesis. Int. J. Oncol. 2014, 45, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-H.; Liu, S.; Leysath, C.E.; Miller-Randolph, S.; Zhang, Y.; Fattah, R.; Bugge, T.H.; Leppla, S.H. Anthrax Toxin Protective Antigen Variants That Selectively Utilize either the CMG2 or TEM8 Receptors for Cellular Uptake and Tumor Targeting. J. Biol. Chem. 2016, 291, 22021–22029. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, J.; Ma, Q.; Cao, L.; Fattah, R.J.; Yu, Z.; Bugge, T.H.; Finkel, T.; Leppla, S.H. Solid tumor therapy by selectively targeting stromal endothelial cells. Proc. Natl. Acad. Sci. USA 2016, 113, E4079–E4087. [Google Scholar] [CrossRef] [PubMed]
- Kappler, M.; Köhler, T.; Kampf, C.; Diestelkötter, P.; Würl, P.; Schmitz, M.; Bartel, F.; Lautenschläger, C.; Rieber, E.P.; Schmidt, H.; et al. Increased survivin transcript levels: An independent negative predictor of survival in soft tissue sarcoma patients. Int. J. Cancer 2001, 95, 360–363. [Google Scholar] [PubMed]
- Würl, P.; Kappler, M.; Meye, A.; Bartel, F.; Köhler, T.; Lautenschläger, C.; Bache, M.; Schmidt, H.; Taubert, H. Co-expression of survivin and TERT and risk of tumour-related death in patients with soft-tissue sarcoma. Lancet 2002, 359, 943–945. [Google Scholar] [CrossRef]
- Szász, A.M.; Lánczky, A.; Nagy, Á.; Förster, S.; Hark, K.; Green, J.E.; Boussioutas, A.; Busuttil, R.; Szabó, A.; Győrffy, B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1065 patients. Oncotarget 2016, 7, 49322–49333. [Google Scholar] [CrossRef] [PubMed]



| Parameter | Description | CMG2 Low Expression (<0.004 fg/fg HPRT) | CMG2 High Expression (>0.004 fg/fg HPRT) | Chi2 Test (p Value) |
|---|---|---|---|---|
| Age | <60 years | 19 | 45 | n.s. |
| >60 years | 11 | 46 | ||
| Gender | female | 11 | 41 | n.s. |
| male | 19 | 50 | ||
| Patient status | alive | 10 | 54 | 0.013 |
| deceased | 20 | 37 | ||
| Tumor stage a | I | 6 | 12 | n.s. |
| II | 8 | 42 | ||
| III | 13 | 28 | ||
| IV | 3 | 9 | ||
| Resection | radical (R0) | 19 | 68 | n.s. |
| not radical (R1) | 11 | 23 | ||
| Tumor localisation | extremities | 19 | 60 | n.s. |
| trunk wall | 3 | 8 | ||
| head/neck | 2 | 2 | ||
| abdomen/retro-peritoneum | 5 | 20 | ||
| multiple locations | 1 | 1 | ||
| Histological subtype | LS | 8 | 23 | n.s. |
| FS | 1 | 7 | ||
| RMS | 3 | 5 | ||
| LMS | 3 | 18 | ||
| NS | 3 | 8 | ||
| Syn | 2 | 5 | ||
| NOS | 6 | 21 | ||
| Other | 4 | 4 | ||
| Tumor size | T1 | 3 | 15 | n.s. |
| T2 | 27 | 76 | ||
| Number of relapses | 0 | 20 | 56 | n.s. |
| 1 | 4 | 19 | ||
| >2 | 6 | 16 |
| Parameter | rs | p | n |
|---|---|---|---|
| CD26 mRNA | 0.26 | 0.005 | 117 |
| uPA mRNA | 0.37 | 0.001 | 73 |
| PAI-1 mRNA | 0.36 | 0.002 | 73 |
| uPAR mRNA | 0.27 | 0.023 | 73 |
| LGR5 mRNA | −0.21 | 0.041 | 92 |
| miR-199a | −0.29 | 0.008 | 85 |
| CMG2 protein | 0.31 | 0.027 | 52 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greither, T.; Wedler, A.; Rot, S.; Keßler, J.; Kehlen, A.; Holzhausen, H.-J.; Bache, M.; Würl, P.; Taubert, H.; Kappler, M. CMG2 Expression Is an Independent Prognostic Factor for Soft Tissue Sarcoma Patients. Int. J. Mol. Sci. 2017, 18, 2648. https://doi.org/10.3390/ijms18122648
Greither T, Wedler A, Rot S, Keßler J, Kehlen A, Holzhausen H-J, Bache M, Würl P, Taubert H, Kappler M. CMG2 Expression Is an Independent Prognostic Factor for Soft Tissue Sarcoma Patients. International Journal of Molecular Sciences. 2017; 18(12):2648. https://doi.org/10.3390/ijms18122648
Chicago/Turabian StyleGreither, Thomas, Alice Wedler, Swetlana Rot, Jacqueline Keßler, Astrid Kehlen, Hans-Jürgen Holzhausen, Matthias Bache, Peter Würl, Helge Taubert, and Matthias Kappler. 2017. "CMG2 Expression Is an Independent Prognostic Factor for Soft Tissue Sarcoma Patients" International Journal of Molecular Sciences 18, no. 12: 2648. https://doi.org/10.3390/ijms18122648