Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards
Abstract
:1. Introduction
2. Results
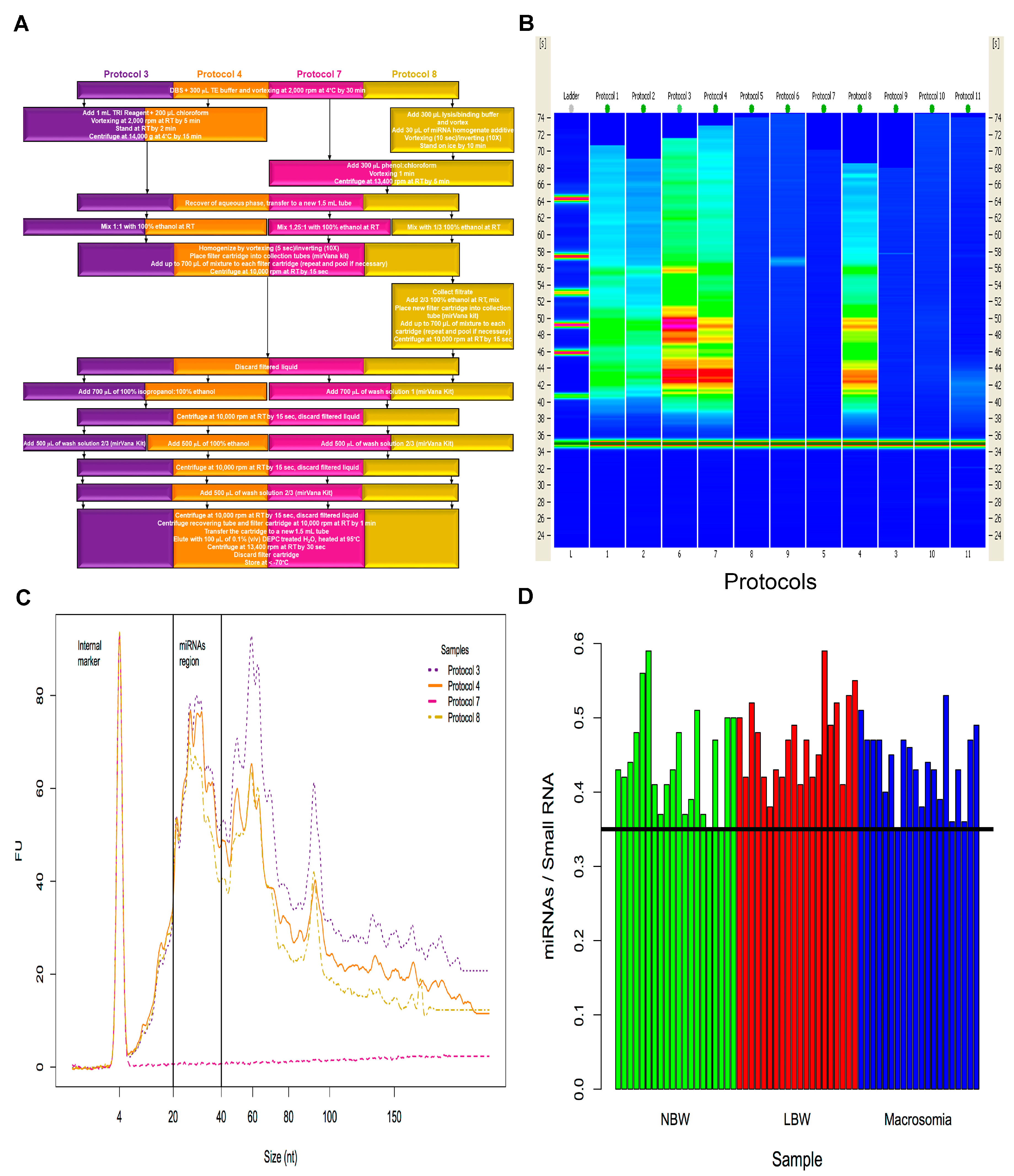
2.1. Isolation of miRNAs from Dried Blood Spots on Newborn Screening Cards (NSC)
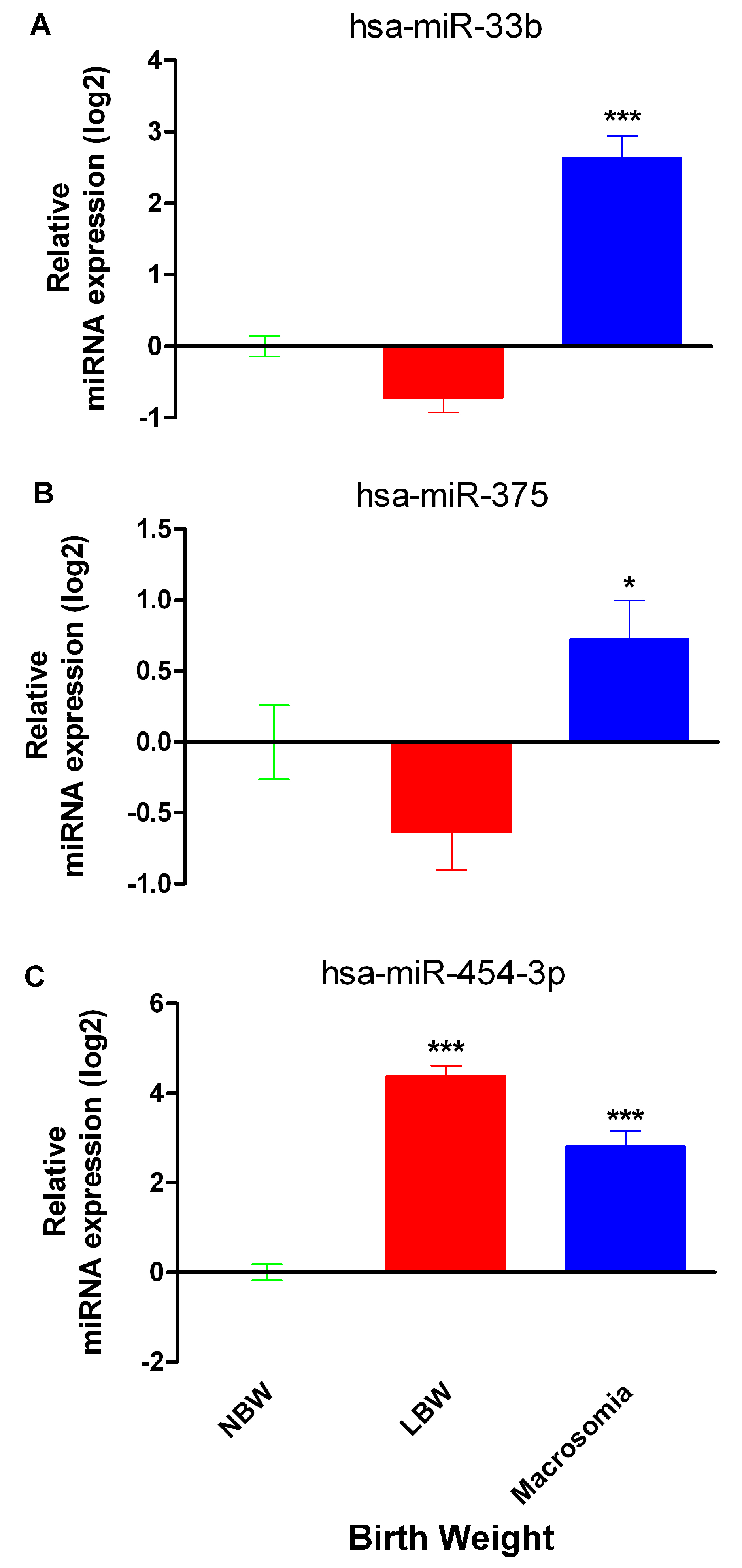
2.2. miR-33b, miR-375 and miR-454-3p Relative Expression in Neonates with Differential Birth Weight
2.3. Prediction of Target Genes and Signaling Pathways for miR-33b, miR-375 and miR-454-3p
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. RNA Purification
4.3. cDNA Synthesis
4.4. Real Time Quantitative PCR
4.5. Metabolic Pathway Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DBS | dried blood spots |
| NSC | newborn screening cards |
| LBW | low birth weight |
| NBW | normal birth weight |
| T2D | type 2 diabetes mellitus |
References
- Langley-Evans, S.C.; McMullen, S. Developmental origins of adult disease. Med. Princ. Pract. 2010, 19, 87–98. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Promoting Optimal Fetal Development: Report of a Technical Consultation. 2006. Available online: http://www.who.int/nutrition/publications/fetomaternal/9241594004/en/ (accessed on 15 July 2014).
- Merzouk, H.; Khan, N.A. Implication of lipids in macrosomia of diabetic pregnancy: Can n-3 polyunsaturated fatty acids exert beneficial effects? Clin. Sci. 2003, 105, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Ramadhani, M.K.; Grobbee, D.E.; Bots, M.L.; Castro-Cabezas, M.; Vos, L.E.; Oren, A.; Uiterwaal, C.S. Lower birth weight predicts metabolic syndrome in young adults: The Atherosclerosis risk in young adults (ARYA)-study. Atherosclerosis 2006, 184, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Gianotti, T.F.; Burgueño, A.L.; Pirola, C.J. Fetal metabolic programming and epigenetic modifications: A systems biology approach. Pediatr. Res. 2013, 73, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Li, J.; Huang, B.; Liu, J.; Chen, X.; Chen, X.M.; Xu, Y.M.; Huang, L.F.; Wang, X.Z. Exosomes: Novel biomarkers for clinical diagnosis. Sci. World J. 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- De Castro, F.; Allen-Leigh, B.; Katz, G.; Salvador-Carulla, L.; Lazcano-Ponce, E. Indicadores de bienestar y desarrollo infantil en México. Salud Pública Mex. 2013, 55, 267–275. [Google Scholar]
- Patnaik, S.K.; Mallick, R.; Yendamuri, S. Detection of microRNAs in dried serum blots. Anal. Biochem. 2010, 407, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Dávalos, A.; Goedeke, L.; Smibert, P.; Ramírez, C.M.; Warrier, N.P.; Andreo, U.; Cirera-Salinas, D.; Rayner, K.; Suresh, U.; Pastor-Pareja, J.C.; et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 9232–9237. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, C.M.; Goedeke, L.; Rotllan, N.; Yoon, J.-H.; Cirera-Salinas, D.; Mattison, J.A.; Suárez, Y.; de Cabo, R.; Gorospe, M.; Fernández-Hernández, C. MicroRNA 33 regulates glucose metabolism. Mol. Cell. Biol. 2013, 33, 2891–2902. [Google Scholar] [CrossRef] [PubMed]
- Goedeke, L.; Vales-Lara, F.M.; Fenstermaker, M.; Cirera-Salinas, D.; Chamorro-Jorganes, A.; Ramírez, C.M.; Mattison, J.A.; de Cabo, R.; Suárez, Y.; Fernández-Hernando, C. A regulatory role for microRNA 33* in controlling lipid metabolism gene expression. Mol. Cell. Biol. 2013, 33, 2339–2352. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Eliasson, L.; Krutzfeldt, J.; Kuwajima, S.; Ma, X.; MacDonald, P.E.; Pfeffer, S.; Tuschl, T.; Rajewsky, N.; Rorsman, P.; et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004, 432, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.Y.; Wen, G.B.; Feng, S.D.; Tuo, Q.-H.; Ou, H.-S.; Yao, C.-H.; Zhu, B.-Y.; Gao, Z.-P.; Zhang, L.; Liao, D.F. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin. Exp. Pharmacol. Physiol. 2011, 38, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Li, X. MiR-375, a microRNA related to diabetes. Gene 2014, 533, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Yu, H.R.; Huang, L.T.; Huang, H.-C.; Chen, R.-F.; Lin, I.-C.; Ou, C.-Y.; Hsu, T.-Y.; Yang, K.D. miRNA-125b regulates TNF-α production in CD14+ neonatal monocytes via post-transcriptional regulation. J. Leukoc. Biol. 2012, 92, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Riffo-Campos, A.L.; Riquelme, I.; Brevi-Meiville, P. Tools for sequence-based miRNA target prediction: What to choose? Int. J. Mol. Sci. 2016, 17, 1987. [Google Scholar] [CrossRef] [PubMed]
- St Julien, K.R.; Jelliffe-Pawlowski, L.L.; Shaw, G.M.; Stevenson, D.K.; O´Brodovich, H.M.; Krasnow, M.A.; Stanford BDP Study Group. High quality genome-wide genotyping from archived dried blood spots without DNA amplification. PLoS ONE 2013, 30. [Google Scholar] [CrossRef]
- Li, Y.; Kowdley, K.V. Method for microRNA isolation from clinical serum samples. Anal. Biochem. 2012, 431, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Moret, I.; Sánchez-Izquierdo, D.; Iborra, M.; Tortosa, L.; Navarro-Puche, A.; Nos, P.; Cervera, J.; Beltrán, B. Assessing an improved protocol for plasma microRNA extraction. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Bergallo, M.; Gambarino, S.; Martino, S.; Montin, D.; Montanari, P.; Galliano, I.; Tovo, P.A. Comparison of two available RNA extraction protocols for microRNA amplification in serum samples. J. Clin. Lab. Anal. 2016, 30, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Czimmerer, Z.; Hulvely, J.; Simandi, Z.; Varallyay, E.; Havelda, Z.; Szabo, E.; Varga, A.; Dezso, B.; Balogh, M.; Horvath, A.; et al. A versatile method to design stem-loop primer-based quantitative PCR assays for detecting small regulatory RNA molecules. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.H.; Zhang, L.H.; Wijesekara, N.; de Haan, W.; Butland, S.; Bhattacharjee, A.; Hayden, M.R. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell. Biol. 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Poy, M.N.; Hausser, J.; Trajkovski, M.; Braun, M.; Collins, S.; Rorsman, P.; Zavolan, M.; Stoffel, M. miR-375 maintains normal pancreatic α- and β-cell mass. Proc. Natl. Acad. Sci. USA 2009, 106, 5813–5818. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Chang, X.; Yin, L.; Li, J.; Zhou, T.; Zhang, C.; Chen, X. Expression and DNA methylation status of microRNA-375 in patients with type 2 diabetes mellitus. Mol. Med. Rep. 2014, 9, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Chang, X.; Li, S.; Li, J.; Yin, L.; Zhou, T.; Zhang, C.; Chen, X.; Sun, K. Ethnic differences in microRNA-375 expression level and DNA methylation status in type 2 diabetes of Han and Kazak populations. J. Diabetes Res. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Somel, M.; Liu, X.; Tang, L.; Yan, Z.; Hu, H.; Guo, S.; Jiang, X.; Xhang, X.; Xu, G.; Xie, G.; et al. MicroRNA-driven developmental remodeling in the brain distinguishes humans from other primates. PLoS Biol. 2011, 9. [Google Scholar] [CrossRef] [PubMed]
- Seyhan, A.A. microRNAs with different functions and roles in disease development and as potential biomarkers of diabetes: Progress and challenges. Mol. Biosyst. 2015, 11, 1217–1234. [Google Scholar] [CrossRef] [PubMed]
- Kitsiou-Tzeli, S.; Tzetis, M. Maternal epigenetics and fetal and neonatal growth. Curr. Opin. Endocrinol. Diabetes Obes. 2017, 1, 43–46. [Google Scholar] [CrossRef] [PubMed]
- American Academy of Pediatrics. Newborn Screening Task Force. Newborn screening: A blueprint for the future. Pediatrics 2000, 106, S383–S427. [Google Scholar]
- Desai, M.; Jellyman, J.K.; Ross, M.G. Epigenomics, gestational programming and risk of metabolic syndrome. Int. J. Obes. 2015, 39, 633–641. [Google Scholar] [CrossRef] [PubMed]
- Zambrano, E. The transgenerational mechanisms in developmental programming of metabolic diseases. Rev. Investig. Clin. 2009, 61, 41–52. [Google Scholar]
- World Health Organization. Child Growth Standards. 2014. Available online: http://www.who.int/childgrowth/standards/weight_for_age_field/en/ (accessed on 15 July 2014).
- George, R.S.; Moat, S.J. Effect of dried blood spot quality on newborn screening analyte concentrations and recommendations for minimum acceptance criteria for sample analysis. Clin. Chem. 2016, 62, 466–475. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viprey, V.F.; Corrias, M.V.; Burchill, S.A. Identification of reference microRNAs and suitability of archived hemopoietic samples for robust microRNA expression profiling. Anal. Biochem. 2012, 421, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, L.; Cheng, K.; Wang, X.; Ren, G.; Xie, P. Identification of suitable plasma-based reference genes for miRNAome analysis of major depressive disorder. J. Affect. Disord. 2014, 163, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Zeng, Y.; Yang, R.; Xu, H.; Chen, Z.; Zhong, J.; Xie, H.; Xu, Y.; Zeng, X. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem. Biophys. Res. Commun. 2014, 454, 210–214. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1–research0034.11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [PubMed]
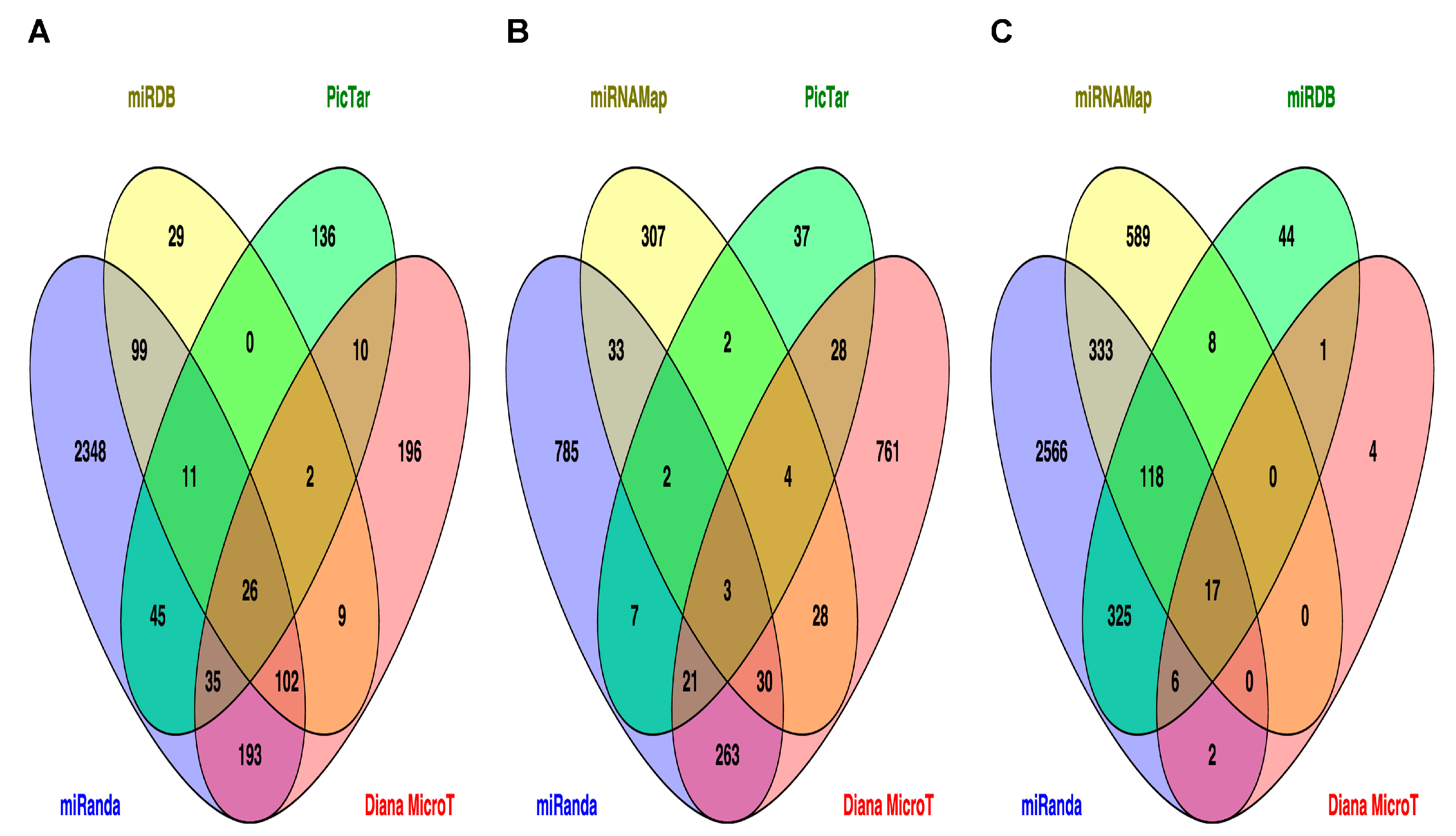
| KEGG Pathway | p Value | Faslse Discovery Rate Adjustment | KEGG ID |
|---|---|---|---|
| Axon guidance | 0.0001975 | 0.00237 | ko04360 |
| cGMP-PKG signaling pathway | 0.01559 | 0.00237 | ko04022 |
| Type 2 diabetes mellitus | 0.01987 | 0.00237 | ko04930 |
| Adherens junction | 0.02716 | 0.00237 | ko04520 |
| GnRH signaling pathway | 0.02716 | 0.04484 | ko04912 |
| Glutamatergic synapse | 0.02894 | 0.04484 | ko04724 |
| Pathogenic Escherichia coli infection | 0.02926 | 0.04484 | ko05130 |
| Cholinergic synapse | 0.03329 | 0.04484 | ko04725 |
| Amphetamine addiction | 0.03363 | 0.04484 | ko05031 |
| Insulin secretion | 0.03774 | 0.04484 | ko04911 |
| Nicotine addiction | 0.0423 | 0.04501 | ko05033 |
| Vascular smooth muscle contraction | 0.04501 | 0.04501 | ko04270 |
| KEGG Pathway | p Value | FDR Adjustment | KEGG ID |
|---|---|---|---|
| Glutamatergic synapse | 0.0108 | 0.03308333 | ko04724 |
| Transcriptional misregulation in cancer | 0.01638 | 0.03308333 | ko05202 |
| Maturity onset diabetes of the young | 0.01985 | 0.03308333 | ko04950 |
| Mitogen-activated protein kinase (MAPK) signaling pathway | 0.03948 | 0.03979 | ko04010 |
| Axon guidance | 0.03979 | 0.03979 | ko04360 |
| KEGG Pathway | p Value | FDR Adjustment | KEGG ID |
|---|---|---|---|
| Endocytosis | 0.001724 | 0.012068 | ko04144 |
| TGF-β signaling pathway | 0.01607 | 0.04421 | ko04350 |
| Axon guidance | 0.02799 | 0.04421 | ko04360 |
| FoxO signaling pathway | 0.03921 | 0.04421 | ko04068 |
| p53 signaling pathway | 0.03971 | 0.04421 | ko04115 |
| Proteoglycans in cancer | 0.04393 | 0.04421 | ko05205 |
| Hippo signaling pathway | 0.04421 | 0.04421 | ko04390 |
| Name | Sequence (5′–3′) | Accession Number of Mature miRNA |
|---|---|---|
| hsa-miR-33b RT stem-loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGCAATG | MIMAT0003301 |
| hsa-miR-33b specific forward | GTTTGGGTGCATTGCTGTTG | |
| hsa-miR-375 RT stem-loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTCACGC | MIMAT0000728 |
| hsa-miR-375 specific forward | TGGTTTTTGTTCGTTCGGCT | |
| hsa-miR-454-3p RT stem-loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCCTA | MIMAT0003885 |
| hsa-miR-454-3p specific forward | GGTGTGGTAGTGCAATATTGCTTA | |
| hsa-miR-106a RT stem-loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCTACCT | MIMAT0000103 |
| hsa-miR-106a specific forward | TGGGTAAAAGTCCTTACAGTGC | |
| hsa-miR-16-5p RT stem-loop | GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCGCCAA | MIMAT0000069 |
| hsa-miR-16-5p specific forward | TGTTTTTTTTTGTAGCAGCACGTAAATA | |
| Universal reverse primer | GTGCAGGGTCCGAGGT | NA |
| Universal ProbeLibrary probe #21 | TGGCTCTG | NA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodil-Garcia, P.; Arellanes-Licea, E.D.C.; Montoya-Contreras, A.; Salazar-Olivo, L.A. Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards. Int. J. Mol. Sci. 2017, 18, 2552. https://doi.org/10.3390/ijms18122552
Rodil-Garcia P, Arellanes-Licea EDC, Montoya-Contreras A, Salazar-Olivo LA. Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards. International Journal of Molecular Sciences. 2017; 18(12):2552. https://doi.org/10.3390/ijms18122552
Chicago/Turabian StyleRodil-Garcia, Patricia, Elvira Del Carmen Arellanes-Licea, Angélica Montoya-Contreras, and Luis A. Salazar-Olivo. 2017. "Analysis of MicroRNA Expression in Newborns with Differential Birth Weight Using Newborn Screening Cards" International Journal of Molecular Sciences 18, no. 12: 2552. https://doi.org/10.3390/ijms18122552





