Nodule-Enriched GRETCHEN HAGEN 3 Enzymes Have Distinct Substrate Specificities and Are Important for Proper Soybean Nodule Development
Abstract
:1. Introduction
2. Results
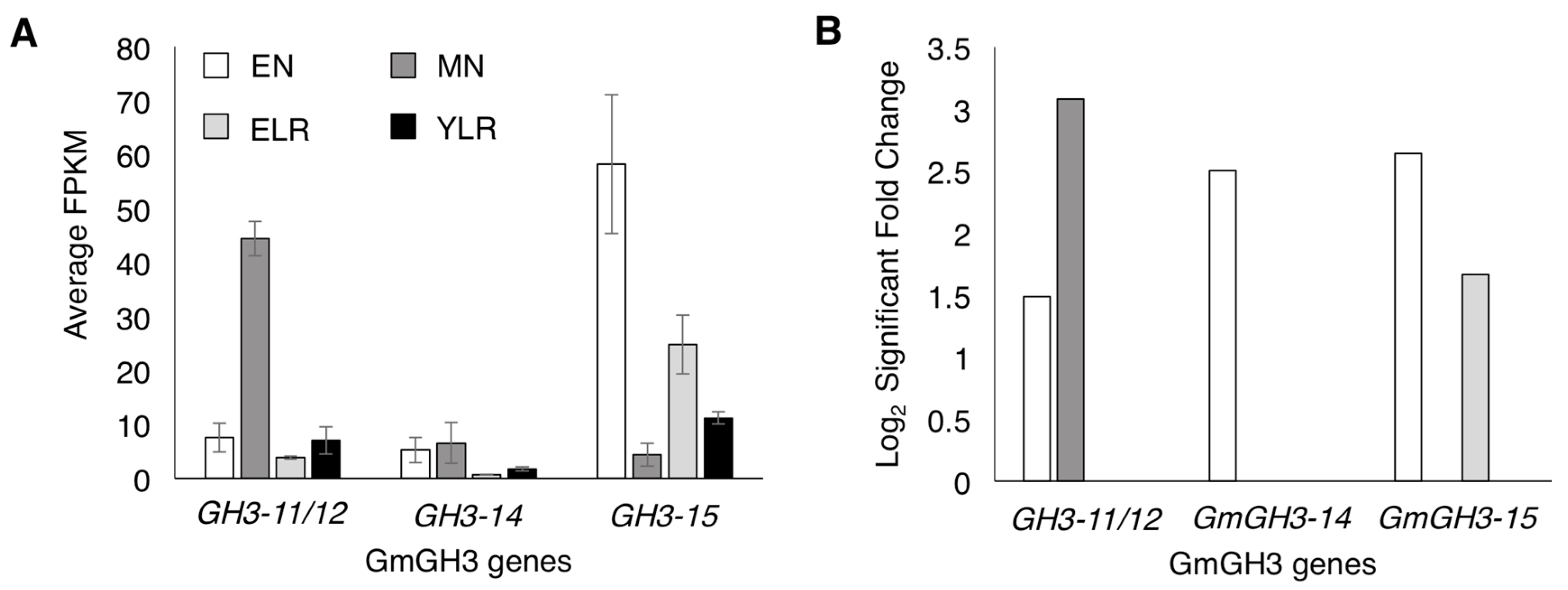
2.1. Identification of Nodule-Enriched GmGH3 Genes
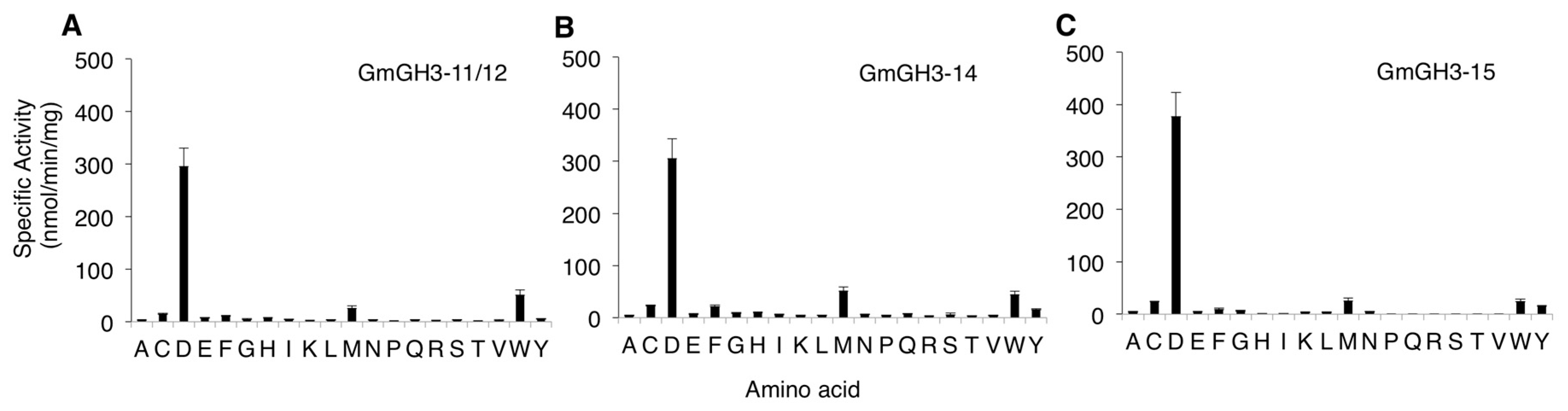
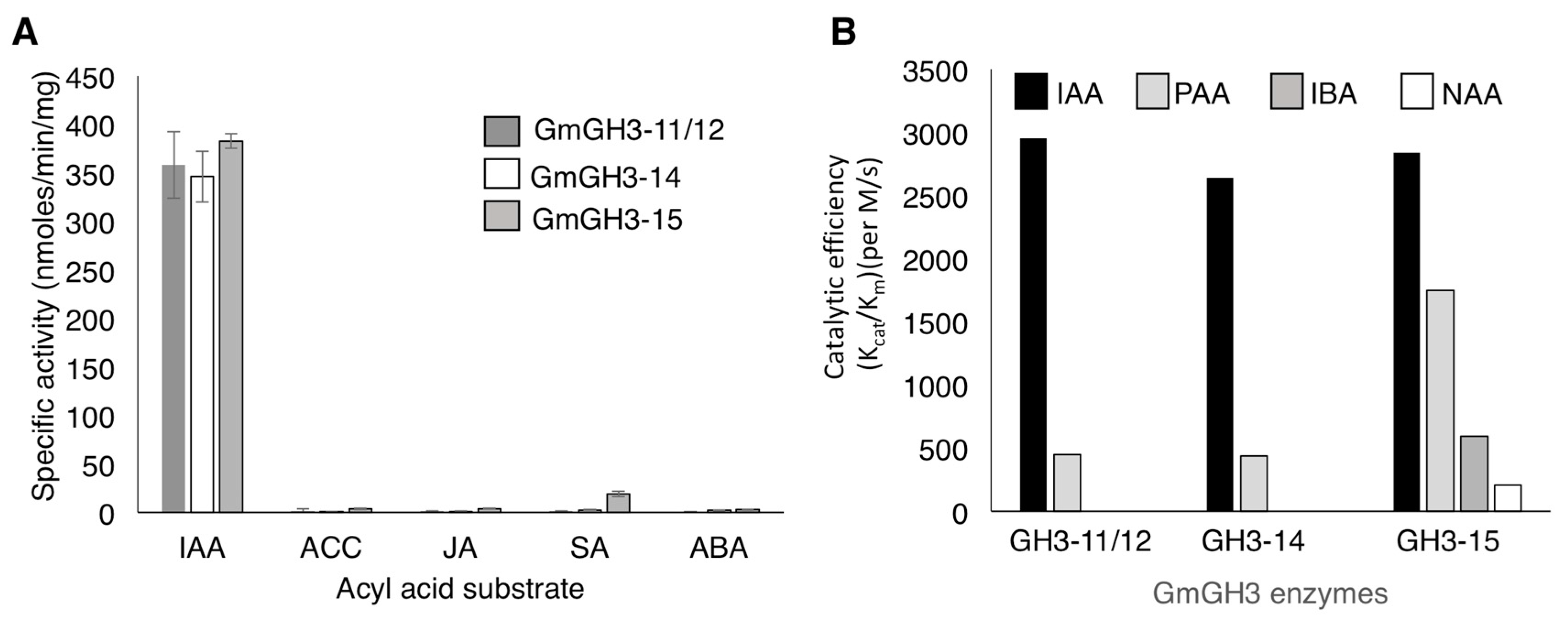
2.2. Nodule-Enriched GmGH3s Show Distinct Acyl Substrate Specificities
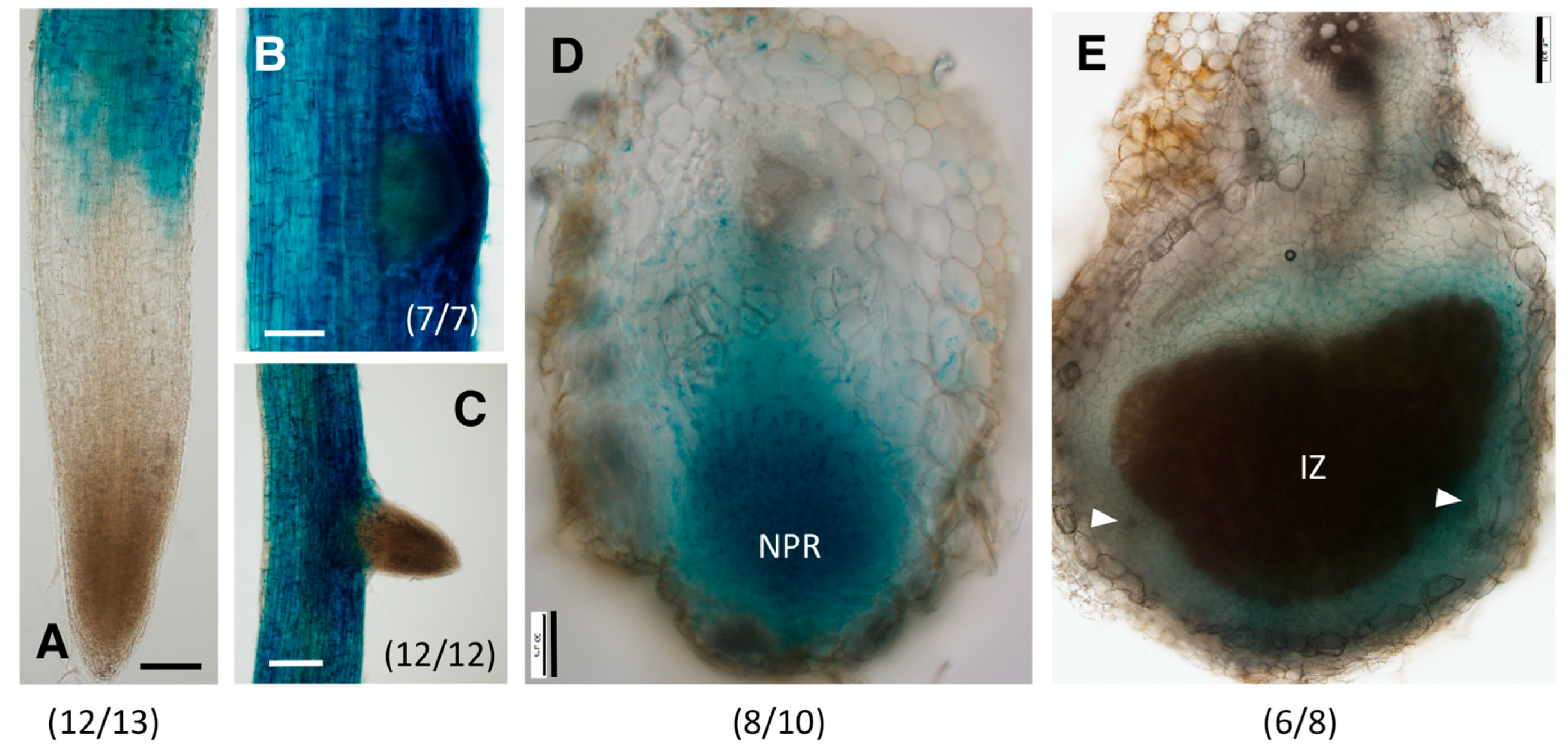
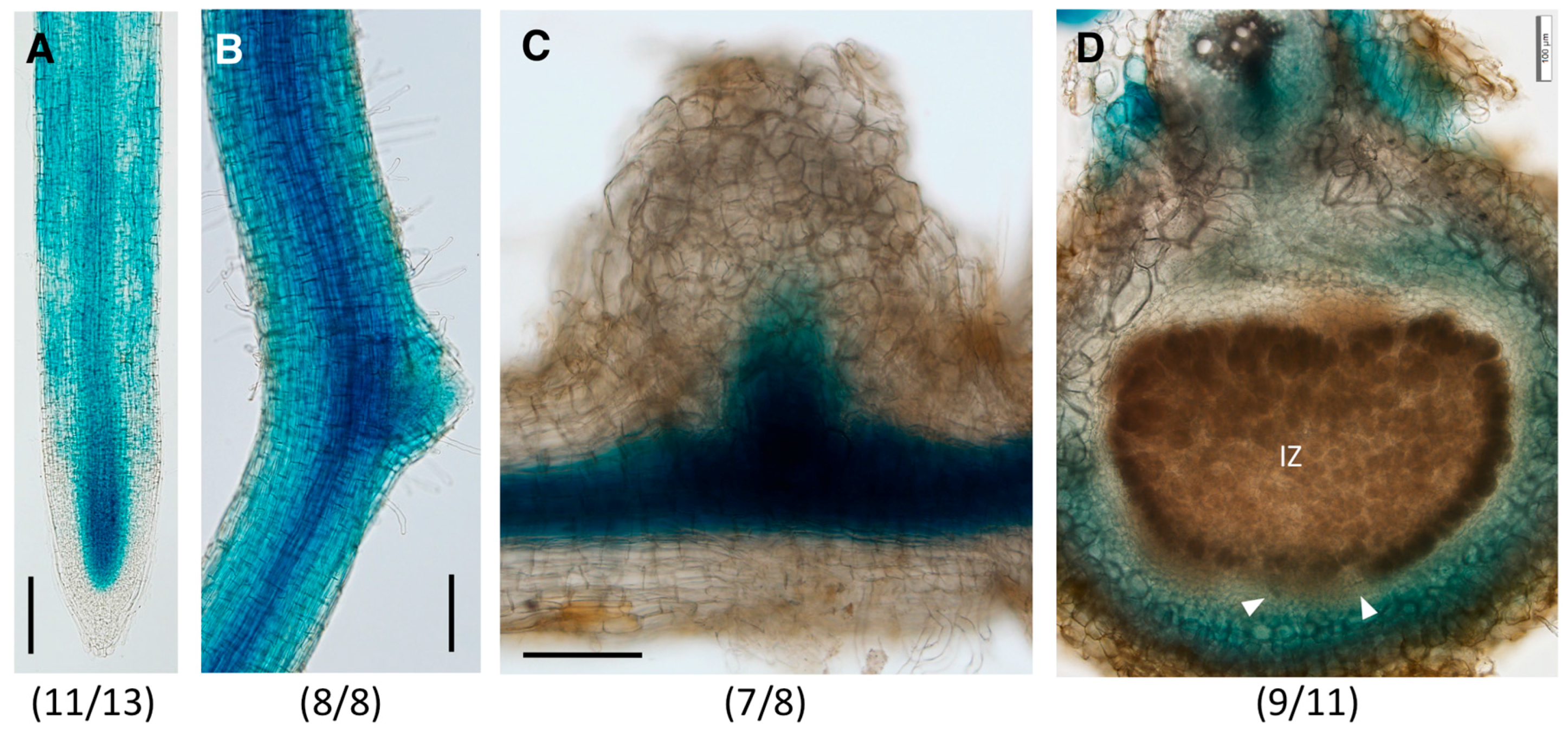
2.3. Distinct Spatio-Temporal Expression Patterns of GmGH3-14 and GmGH3-15 in Soybean Roots and Nodules
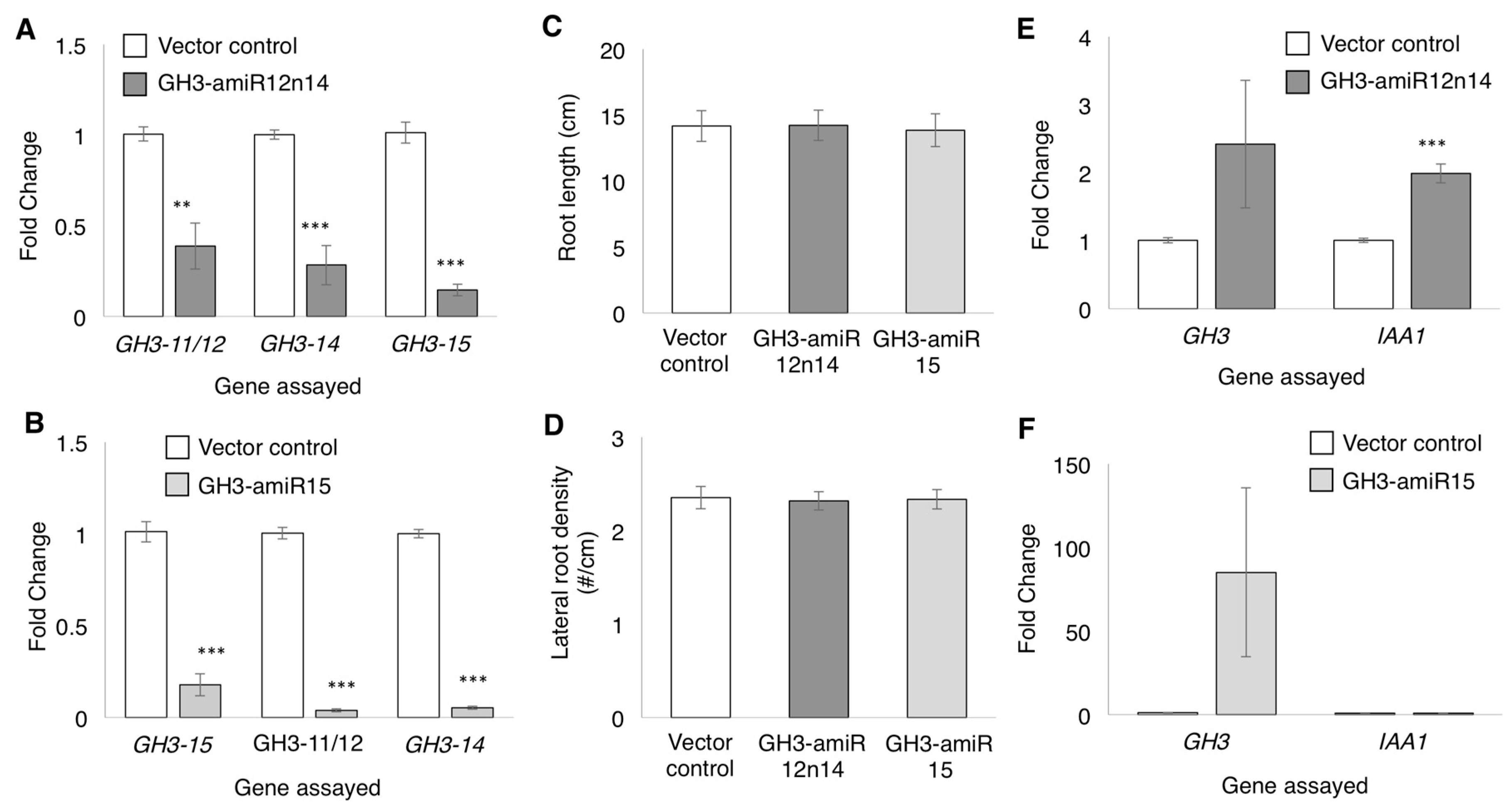
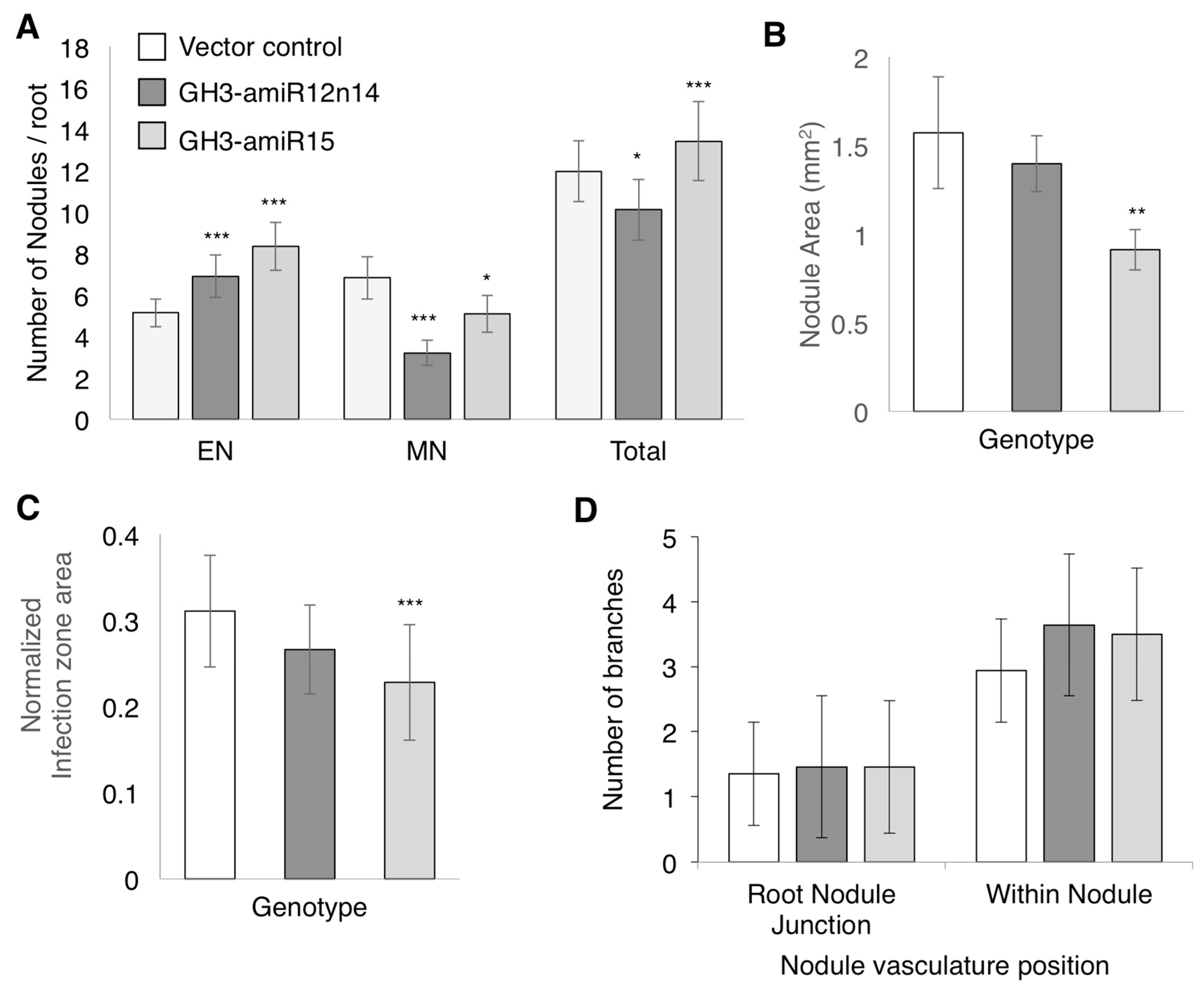
2.4. GmGH3-14 and GmGH3-15 Are Important for Proper Nodule Numbers in Soybean
2.5. GmGH3s Influence Nodule Size in Soybean
3. Discussion
4. Materials and Methods
4.1. Protein Expression, Purification, and Enzyme Assays
4.2. Cloning for Promoter:GUS and Artificial miRNA
4.3. Plant Material and Growth Conditions
4.4. Plant Transformation and Nodulation Assay
4.5. GUS Staining and Microscopy
4.6. Phloroglucinol Staining
4.7. Quantitative Gene Expression Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GH3 | Gretchen Hagen 3 |
| IAA | indole-3-acetic acid |
| IPA | indole pyruvic acid |
| oxIAA | 2-oxoindole-3-acetic acid |
| ARF | auxin response factor |
| ACC | 1-aminocyclopropane carboxylic acid |
| ABA | abscisic acid |
| JA | jasmonic acid |
| SA | salicylic acid |
| PAA | phenyl acetic acid |
| IBA | indole butyric acid |
| NAA | naphthalene acetic acid |
| GUS | β-glucuronidase |
| RT-qPCR | reverse transcription-quantitative polymerase chain reaction |
| BA | benzoic acid |
| 4-HBA | 4-hydorxy benzoic acid |
| N− PNS | nitrogen-free plant nutrient solution |
References
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Herrmann, J.; Chen, Q.; Wang, S.; Jez, J.M. Modulating plant hormones by enzyme action: The GH3 family of acyl acid amido synthetases. Plant Signal. Behav. 2010, 5, 1607–1612. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin Biosynthesis: A Simple Two-Step Pathway Converts Tryptophan to Indole-3-Acetic Acid in Plants. Mol. Plant 2012, 5, 334–338. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Dolezal, K.; Schlereth, A.; Jurgens, G.; Alonso, J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Pencik, A.; Simonovik, B.; Petersson, S.V.; Henykova, E.; Simon, S.; Greenham, K.; Zhang, Y.; Kowalczyk, M.; Estelle, M.; Zazimalova, E.; et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 2013, 25, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Cheng, Y.; Murphy, A.S. Evidence of oxidative attenuation of auxin signalling. J. Exp. Bot. 2013, 64, 2629–2639. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, Y.; Liu, X.; Zhang, X.; Liu, S.; Yu, X.; Ren, Y.; Zheng, X.; Zhou, K.; Jiang, L.; et al. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 2013, 27, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [PubMed]
- Porco, S.; Pencik, A.; Rashed, A.; Voss, U.; Casanova-Saez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Muller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Korasick, D.A.; Enders, T.A.; Strader, L.C. Auxin biosynthesis and storage forms. J. Exp. Bot. 2013, 64, 2541–2555. [Google Scholar] [CrossRef] [PubMed]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [PubMed]
- LeClere, S.; Tellez, R.; Rampey, R.A.; Matsuda, S.P.; Bartel, B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002, 277, 20446–20452. [Google Scholar] [CrossRef] [PubMed]
- Rampey, R.A.; LeClere, S.; Kowalczyk, M.; Ljung, K.; Sandberg, G.; Bartel, B. A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 2004, 135, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T.J. Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 1985, 5, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-W.; Li, C.-H.; Cao, J.; Zhang, Y.-C.; Zhang, S.-Q.; Xia, Y.-F.; Sun, D.-Y.; Sun, Y. Altered Architecture and Enhanced Drought Tolerance in Rice via the Down-Regulation of Indole-3-Acetic Acid by TLD1/OsGH3.13 Activation. Plant Physiol. 2009, 151, 1889–1901. [Google Scholar] [CrossRef] [PubMed]
- Böttcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of auxin by the indole-3-acetic acid-amido synthetase GH3–1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Park, J.Y.; Kim, Y.S.; Staswick, P.E.; Jeon, J.; Yun, J.; Kim, S.Y.; Kim, J.; Lee, Y.H.; Park, C.M. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 2007, 282, 10036–10046. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Guo, Y.; Novák, O.; Chen, W.; Ljung, K.; Noel, J.P.; Chory, J. Local auxin metabolism regulates environment-induced hypocotyl elongation. Nat. Plants 2016, 2, 16025. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Nakazawa, M.; Ishikawa, A.; Kawashima, M.; Ichikawa, T.; Takahashi, N.; Shimada, H.; Manabe, K.; Matsui, M. ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J. 2004, 37, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Stone, J.M. Arabidopsis thaliana GH3.9 influences primary root growth. Planta 2007, 226, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Zubieta, C.; Herrmann, J.; Kapp, U.; Nanao, M.H.; Jez, J.M. Structural basis for prereceptor modulation of plant hormones by GH3 proteins. Science 2012, 336, 1708–1711. [Google Scholar] [CrossRef] [PubMed]
- Peat, T.S.; Bottcher, C.; Newman, J.; Lucent, D.; Cowieson, N.; Davies, C. Crystal structure of an indole-3-acetic acid amido synthetase from grapevine involved in auxin homeostasis. Plant Cell 2012, 24, 4525–4538. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis Enzyme Family That Conjugates Amino Acids to Indole-3-Acetic Acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Breakspear, A.; Liu, C.; Roy, S.; Stacey, N.; Rogers, C.; Trick, M.; Morieri, G.; Mysore, K.S.; Wen, J.; Oldroyd, G.E.; et al. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 2014, 26, 4680–4701. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Wang, Y.; Zhu, L.; Tian, Y.; Chen, L.; Sun, Z.; Ullah, I.; Li, X. GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 2017, 215, 672–686. [Google Scholar] [CrossRef] [PubMed]
- Turner, M.; Nizampatnam, N.R.; Baron, M.; Coppin, S.; Damodaran, S.; Adhikari, S.; Arunachalam, S.P.; Yu, O.; Subramanian, S. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013, 162, 2042–2055. [Google Scholar] [CrossRef] [PubMed]
- Nizampatnam, N.R.; Schreier, S.J.; Damodaran, S.; Adhikari, S.; Subramanian, S. microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015, 84, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, K.; Chen, L.; Zou, Y.; Liu, H.; Tian, Y.; Li, D.; Wang, R.; Zhao, F.; Ferguson, B.J.; et al. MicroRNA167-Directed Regulation of the Auxin Response Factors GmARF8a and GmARF8b Is Required for Soybean Nodulation and Lateral Root Development. Plant Physiol. 2015, 168, 984–999. [Google Scholar] [CrossRef] [PubMed]
- Hobecker, K.V.; Reynoso, M.A.; Bustos Sanmamed, P.; Wen, J.; Mysore, K.S.; Crespi, M.; Blanco, F.A.; Zanetti, M.E. The microRNA390/TAS3 pathway mediates in symbiotic nodulation and lateral root growth. Plant Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Ng, J.L.P.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signalling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2017, erx308. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, A.M. Developmental biology of legume nodulation. New Phytol. 1992, 122, 211–237. [Google Scholar] [CrossRef]
- Sprent, J.I.; James, E.K. Legume Evolution: Where Do Nodules and Mycorrhizas Fit in? Plant Physiol. 2007, 144, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Mathesius, U.; Schlaman, H.R.; Spaink, H.P.; Of Sautter, C.; Rolfe, B.G.; Djordjevic, M.A. Auxin transport inhibition precedes root nodule formation in white clover roots and is regulated by flavonoids and derivatives of chitin oligosaccharides. Plant J. 1998, 14, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Pacios-Bras, C.; Schlaman, H.R.M.; Boot, K.; Admiraal, P.; Mateos Langerak, J.; Stougaard, J.; Spaink, H.P. Auxin distribution in Lotus japonicus during root nodule development. Plant Mol. Biol. 2003, 52, 1169–1180. [Google Scholar] [CrossRef] [PubMed]
- Takanashi, K.; Sugiyama, A.; Yazaki, K. Involvement of auxin distribution in root nodule development of Lotus japonicus. Planta 2011, 234, 73–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzaki, T.; Yano, K.; Ito, M.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 2012, 139, 3997–4006. [Google Scholar] [CrossRef] [PubMed]
- Wasson, A.P.; Pellerone, F.I.; Mathesius, U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell 2006, 18, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Schnabel, E.; Hughes, K.; Frugoli, J. RNAi Phenotypes and the Localization of a Protein::GUS Fusion Imply a Role for Medicago truncatula PIN Genes in Nodulation. J. Plant Growth Regul. 2006, 25, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Sanko-Sawczenko, I.; Lotocka, B.; Czarnocka, W. Expression Analysis of PIN Genes in Root Tips and Nodules of Medicago truncatula. Int. J. Mol. Sci. 2016, 17, 1197. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Stacey, G.; Yu, O. Endogenous isoflavones are essential for the establishment of symbiosis between soybean and Bradyrhizobium japonicum. Plant J. 2006, 48, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Mathesius, U. Flavonoids induced in cells undergoing nodule organogenesis in white clover are regulators of auxin breakdown by peroxidase. J. Exp. Bot. 2001, 52, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Damodaran, S.; Adhikari, S.; Lin, J.; Pathak, S.; Ge, X.; Blakeslee, L.; Subramanian, S. Comparative transcriptomics reveals a role for GmCYP83B1 in regulating auxin homeostasis during soybean nodule development. Plant Physiol. 2017. submitted. [Google Scholar]
- Yang, Y.; Yue, R.; Sun, T.; Zhang, L.; Chen, W.; Zeng, H.; Wang, H.; Shen, C. Genome-wide identification, expression analysis of GH3 family genes in Medicago truncatula under stress-related hormones and Sinorhizobium meliloti infection. Appl. Microbiol. Biotechnol. 2015, 99, 841–854. [Google Scholar] [CrossRef] [PubMed]
- Campanella, J.J.; Smith, S.M.; Leibu, D.; Wexler, S.; Ludwig-Müller, J. The Auxin Conjugate Hydrolase Family of Medicago truncatula and Their Expression During the Interaction with Two Symbionts. J. Plant Growth Regul. 2008, 27, 26–38. [Google Scholar] [CrossRef]
- Fukuhara, H.; Minakawa, Y.; Akao, S.; Minamisawa, K. The Involvement of Indole-3-Acetic Acid Produced by Bradyrhizobium elkanii in Nodule Formation. Plant Cell Physiol. 1994, 35, 1261–1265. [Google Scholar] [CrossRef]
- Perrine, F.M.; Rolfe, B.G.; Hynes, M.F.; Hocart, C.H. Gas chromatography-mass spectrometry analysis of indoleacetic acid and tryptophan following aqueous chloroformate derivatisation of Rhizobium exudates. Plant Physiol. Biochem. PPB 2004, 42, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Petrasek, J. Why plants need more than one type of auxin. Plant Sci. 2011, 180, 454–460. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly Specific Gene Silencing by Artificial MicroRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Elmore, J.M.; Fester, T.; Taylor, C.G. Evaluation of constitutive viral promoters in transgenic soybean roots and nodules. Mol. Plant-Microbe Interact. 2008, 21, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Westfall, C.S.; Sherp, A.M.; Zubieta, C.; Alvarez, S.; Schraft, E.; Marcellin, R.; Ramirez, L.; Jez, J.M. Arabidopsis thaliana GH3.5 acyl acid amido synthetase mediates metabolic crosstalk in auxin and salicylic acid homeostasis. Proc. Natl. Acad. Sci. USA 2016, 113, 13917–13922. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dai, X.; Liu, T.; Zhao, P.X. LegumeIP: An integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 2012, 40, D1221–D1229. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, S.; Mashiguchi, K.; Tanaka, K.; Hishiyama, S.; Sakai, T.; Hanada, K.; Kinoshita-Tsujimura, K.; Yu, H.; Dai, X.; Takebayashi, Y.; et al. Distinct Characteristics of Indole-3-Acetic Acid and Phenylacetic Acid, Two Common Auxins in Plants. Plant Cell Physiol. 2015, 56, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Q.; Li, Z.; Staswick, P.E.; Wang, M.; Zhu, Y.; He, Z. Dual Regulation Role of GH3.5 in Salicylic Acid and Auxin Signaling during Arabidopsis-Pseudomonas syringae Interaction. Plant Physiol. 2007, 145, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Van Spronsen, P.C.; Tak, T.; Rood, A.M.; van Brussel, A.A.; Kijne, J.W.; Boot, K.J. Salicylic acid inhibits indeterminate-type nodulation but not determinate-type nodulation. Mol. Plant-Microbe Interact. 2003, 16, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Stacey, G.; McAlvin, C.B.; Kim, S.Y.; Olivares, J.; Soto, M.J. Effects of endogenous salicylic acid on nodulation in the model legumes Lotus japonicus and Medicago truncatula. Plant Physiol. 2006, 141, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Hammad, Y.; Nalin, R.; Marechal, J.; Fiasson, K.; Pepin, R.; Berry, A.M.; Normand, P.; Domenach, A.-M. A possible role for phenyl acetic acid (PAA) on Alnus glutinosa nodulation by Frankia. Plant Soil 2003, 254, 193–205. [Google Scholar] [CrossRef]
- Chen, Q.; Westfall, C.S.; Hicks, L.M.; Wang, S.; Jez, J.M. Kinetic Basis for the Conjugation of Auxin by a GH3 Family Indole-acetic Acid-Amido Synthetase. J. Biol. Chem. 2010, 285, 29780–29786. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.; Fuchs, B.; Walter, N.; Kevin Lutke, W.; Taylor, C.G. Ex vitro composite plants: An inexpensive, rapid method for root biology. Plant J. 2005, 43, 449–457. [Google Scholar] [CrossRef] [PubMed]
- Bhuvaneswari, T.V.; Turgeon, B.G.; Bauer, W.D. Early Events in the Infection of Soybean (Glycine max L. Merr) by Rhizobium japonicum: I. Localization of infectible root cells. Plant Physiol. 1980, 66, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Libault, M.; Thibivilliers, S.; Bilgin, D.D.; Radwan, O.; Benitez, M.; Clough, S.J.; Stacey, G. Identification of Four Soybean Reference Genes for Gene Expression Normalization. Plant Genome 2008, 1, 44–54. [Google Scholar] [CrossRef]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damodaran, S.; Westfall, C.S.; Kisely, B.A.; Jez, J.M.; Subramanian, S. Nodule-Enriched GRETCHEN HAGEN 3 Enzymes Have Distinct Substrate Specificities and Are Important for Proper Soybean Nodule Development. Int. J. Mol. Sci. 2017, 18, 2547. https://doi.org/10.3390/ijms18122547
Damodaran S, Westfall CS, Kisely BA, Jez JM, Subramanian S. Nodule-Enriched GRETCHEN HAGEN 3 Enzymes Have Distinct Substrate Specificities and Are Important for Proper Soybean Nodule Development. International Journal of Molecular Sciences. 2017; 18(12):2547. https://doi.org/10.3390/ijms18122547
Chicago/Turabian StyleDamodaran, Suresh, Corey S. Westfall, Brian A. Kisely, Joseph M. Jez, and Senthil Subramanian. 2017. "Nodule-Enriched GRETCHEN HAGEN 3 Enzymes Have Distinct Substrate Specificities and Are Important for Proper Soybean Nodule Development" International Journal of Molecular Sciences 18, no. 12: 2547. https://doi.org/10.3390/ijms18122547





