Regulation of the Tumor-Suppressor BECLIN 1 by Distinct Ubiquitination Cascades
Abstract
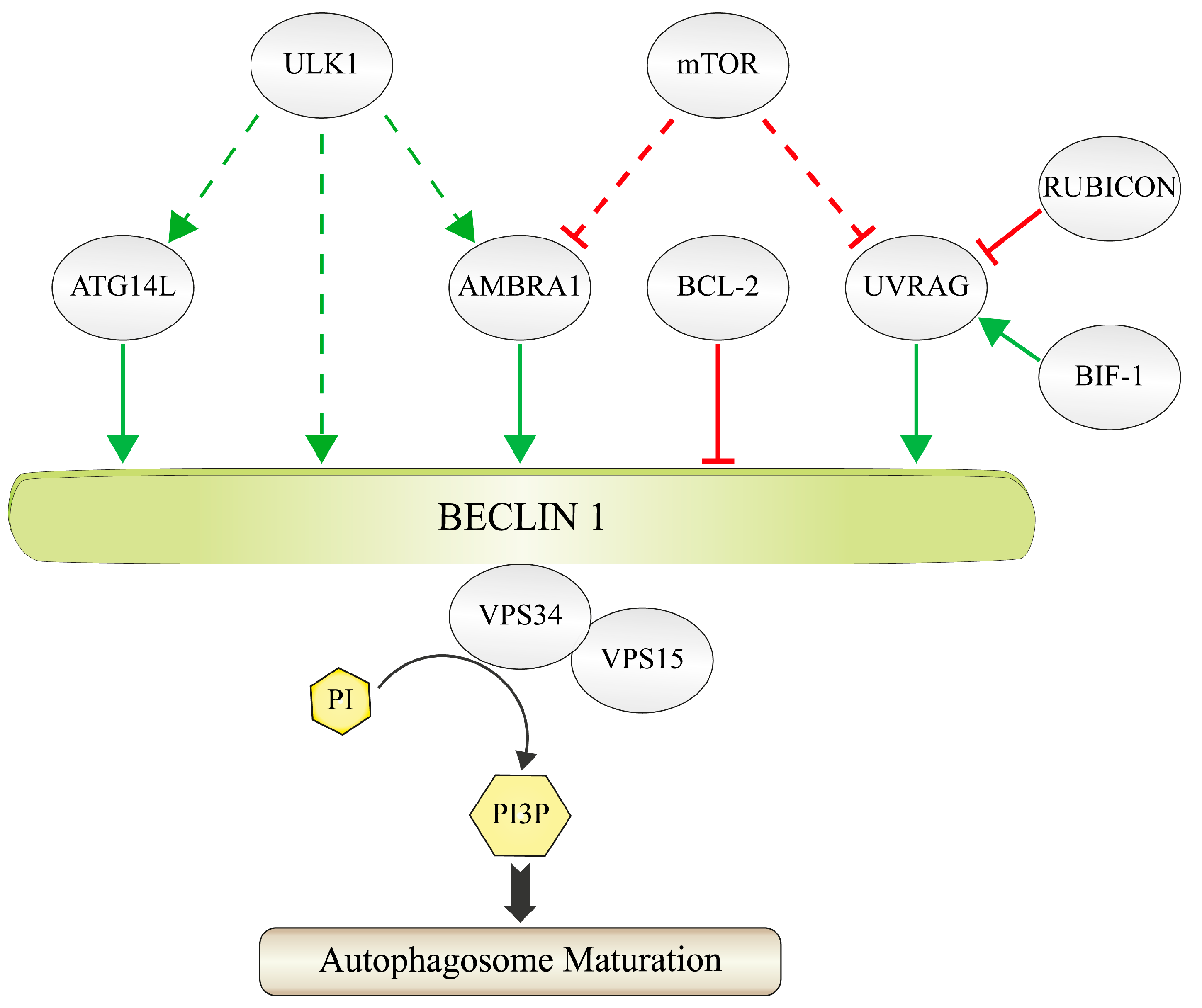
:1. Formation and Function of the PI3K-III Complex
1.1. Principles of PtdIns3P-Signaling during Autophagy
1.2. The Composition of the PI3K-III Complex
2. Regulation of BECLIN 1 via Distinct Ubiquitination Machineries
2.1. Lys63-Linked Polyubiquitination of BECLIN 1 by the AMBRA 1-Containing CUL4-Ligase Complex
2.2. Lys63-Linked Polyubiquitination and Activation of BECLIN 1 by TRAF6 during Phagocytosis
2.3. USP14 Suppresses Autophagy by Cleaving Lys63-Ubiquitin Chains from BECLIN 1
2.4. Lys11-Linked Polyubiquitination and Degradation of BECLIN 1 by NEDD4
2.5. Deubiquitination of Lys11-Linked Polyubiquitin Chains and Stabilization of BECLIN 1 by USP19 and USP13
2.6. Chemical Inhibition of the Deubiquitinating Enzymes USP10 and USP13 Causes Ubiquitination and Degradation of BECLIN 1
2.7. RNF216 Catalyzes Lys48-Linked Polyubiquitination of BECLIN 1 and Marks it for Degradation
2.8. USP9X Governs the Reciprocal Ubiquitin-Based Interplay of BECLIN 1 and MCL-1
2.9. The Chaperone HSP90 Protects BECLIN 1 against Lys48-Linked Polyubiquitination and Degradation
2.10. Limitation of Autophagy by KLHL20-Catalyzed Lys48-Linked Ubiquitination of BECLIN 1, ULK1 and VPS34
2.11. Competition with the Ubiquitin-Like ISG15 Interferes with Polyubiquitin Chain Formation on BECLIN 1
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Avalos, Y.; Canales, J.; Bravo-Sagua, R.; Criollo, A.; Lavandero, S.; Quest, A.F. Tumor Suppression and Promotion by Autophagy. Biomed. Res. Int. 2014. [Google Scholar] [CrossRef]
- Lozy, F.; Karantza, V. Autophagy and cancer cell metabolism. Semin. Cell. Dev. Biol. 2012, 23, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Brech, A.; Ahlquist, T.; Lothe, R.A.; Stenmark, H. Autophagy in tumour suppression and promotion. Mol. Oncol. 2009, 3, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Yin, Z.; Pascual, C.; Klionsky, D.J. Autophagy: Machinery and regulation. Microb. Cell 2016, 3, 588–596. [Google Scholar] [CrossRef] [PubMed]
- Farrell, F.; Rusten, T.E.; Stenmark, H. Phosphoinositide 3-kinases as accelerators and brakes of autophagy. FEBS J. 2013, 280, 6322–6337. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Guillermet-Guibert, J.; Graupera, M.; Bilanges, B. The emerging mechanisms of isoform-specific PI3K signalling. Nat. Rev. Mol. Cell. Biol. 2010, 11, 329–341. [Google Scholar] [CrossRef] [PubMed]
- De Craene, J.O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, E634. [Google Scholar] [CrossRef] [PubMed]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127, 923–928. [Google Scholar] [CrossRef] [PubMed]
- Backer, J.M. The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochem. J. 2008, 410, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Schu, P.V.; Takegawa, K.; Fry, M.J.; Stack, J.H.; Waterfield, M.D.; Emr, S.D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 1993, 260, 88–91. [Google Scholar] [CrossRef] [PubMed]
- Workman, P.; van Montfort, R.L. Unveiling the secrets of the ancestral PI3 kinase Vps34. Cancer Cell 2010, 17, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Backer, J.M. The intricate regulation and complex functions of the Class III phosphoinositide 3-kinase Vps34. Biochem. J. 2016, 473, 2251–2271. [Google Scholar] [CrossRef] [PubMed]
- Reidick, C.; Boutouja, F.; Platta, H.W. The class III phosphatidylinositol 3-kinase Vps34 in Saccharomyces cerevisiae. Biol. Chem. 2017, 398, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Welters, P.; Takegawa, K.; Emr, S.D.; Chrispeels, M.J. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc. Natl. Acad. Sci. USA 1994, 91, 11398–11402. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef] [PubMed]
- Ellson, C.D.; Anderson, K.E.; Morgan, G.; Chilvers, E.R.; Lipp, P.; Stephens, L.R.; Hawkins, P.T. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr. Biol. 2001, 11, 1631–1635. [Google Scholar] [CrossRef]
- Vieira, O.V.; Botelho, R.J.; Rameh, L.; Brachmann, S.M.; Matsuo, T.; Davidson, H.W.; Schreiber, A.; Backer, J.M.; Cantley, L.C.; Grinstein, S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 2001, 155, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Sagona, A.P.; Nezis, I.P.; Pedersen, N.M.; Liestøl, K.; Poulton, J.; Rusten, T.E.; Skotheim, R.I.; Raiborg, C.; Stenmark, H. PtdIns(3)P controls cytokinesis through KIF13A-mediated recruitment of FYVE-CENT to the midbody. Nat. Cell Biol. 2010, 12, 362–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunau, S.; Lay, D.; Mindthoff, S.; Platta, H.W.; Girzalsky, W.; Just, W.W.; Erdmann, R. The Phosphoinositide-3-kinase Vps34p is required for pexophagy in Saccharomyces cerevisae. Biochem. J. 2011, 434, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Hayashi-Nishino, M.; Fujita, N.; Noda, T.; Yamaguchi, A.; Yoshimori, T.; Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009, 11, 1433–1437. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.S.; Zhang, H.; Prinz, W.A. Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 2017, 19, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9, 99–111. [Google Scholar] [CrossRef] [PubMed]
- Funderburk, S.F.; Wang, Q.J.; Yue, Z. The Beclin 1-VPS34 complex—At the crossroads of autophagy and beyond. Trends Cell Biol. 2010, 20, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Nezis, I.P.; Sagona, A.P.; Schink, K.O.; Stenmark, H. Divide and ProsPer: The emerging role of PtdIns3P in cytokinesis. Trends Cell Biol. 2010, 20, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Stenmark, H. Endocytosis and signaling. Curr. Opin. Cell Biol. 2011, 23, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, A.; Tooze, S.A. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009, 186, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Stack, J.H.; Herman, P.K.; Schu, P.V.; Emr, S.D. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993, 12, 2195–2204. [Google Scholar] [PubMed]
- He, C.; Levine, B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010, 22, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Abrahamsen, H.; Stenmark, H.; Platta, H.W. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: Tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett. 2012, 586, 1584–1591. [Google Scholar] [CrossRef] [PubMed]
- El Magraoui, E.; Reidick, C.; Meyer, H.E.; Platta, H.W. Autophagy-Related Deubiquitinating Enzymes Involved in Health and Disease. Cells 2015, 4, 596–621. [Google Scholar] [CrossRef] [PubMed]
- Kuang, E.; Qi, J.; Ronai, Z. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem. Sci. 2013, 38, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Reidick, C.; El Magraoui, F.; Meyer, H.E.; Stenmark, H.; Platta, H.W. Regulation of the Tumor-Suppressor Function of the Class III Phosphatidylinositol 3-Kinase Complex by Ubiquitin and SUMO. Cancers 2015, 7, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, M.; Di Rienzo, M.; Piacentini, M.; Fimia, G.M. Emerging Mechanisms in Initiating and Terminating Autophagy. Trends Biochem. Sci. 2017, 42, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Itakura, E.; Kishi, C.; Inoue, K.; Mizushima, N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 2008, 19, 5360–53672. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Fan, W.; Chen, K.; Ding, X.; Chen, S.; Zhong, Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 2008, 105, 19211–19226. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Wang, Q.J.; Li, X.; Yan, Y.; Backer, J.M.; Chait, B.T.; Heintz, N.; Yue, Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 2009, 11, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Saitoh, T.; Tabata, K.; Omori, H.; Satoh, T.; Kurotori, N.; Maejima, I.; Shirahama-Noda, K.; Ichimura, T.; Isobe, T.; Akira, S.; Noda, T.; Yoshimori, T. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 2009, 11, 385–396. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, K.; Morita, E.; Saitoh, T.; Akira, S.; Ktistakis, N.T.; Izumi, T.; Noda, T.; Yoshimori, T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 2010, 190, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; Amano, A.; Yoshimori, T. Autophagosomes form at ER-mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Nassiri, A.; Zhong, Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc. Natl. Acad. Sci. USA 2011, 108, 7769–7774. [Google Scholar] [CrossRef] [PubMed]
- Fogel, A.I.; Dlouhy, B.J.; Wang, C.; Ryu, S.W.; Neutzner, A.; Hasson, S.A.; Sideris, D.P.; Abeliovich, H.; Youle, R.J. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol. 2013, 33, 3675–3688. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Zhong, Q.; Sheng, Z.H.; Yoshimori, T.; Liang, C.; Jung, J.U. Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J. Cell Sci. 2012, 125, 4740–4750. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Stoykova, A.; Romagnoli, A.; Giunta, L.; Di Bartolomeo, S.; Nardacci, R.; Corazzari, M.; Fuoco, C.; Ucar, A.; Schwartz, P.; et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007, 447, 1121–1125. [Google Scholar] [PubMed]
- Di Bartolomeo, S.; Corazzari, M.; Nazio, F.; Oliverio, S.; Lisi, G.; Antonioli, M.; Pagliarini, V.; Matteoni, S.; Fuoco, C.; Giunta, L.; et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 2010, 191, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Fimia, G.M.; Corazzari, M.; Antonioli, M.; Piacentini, M. Ambra1 at the crossroad between autophagy and cell death. Oncogene 2013, 32, 3311–3318. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Lee, J.S.; Inn, K.S.; Gack, M.U.; Li, Q.; Roberts, E.A.; Vergne, I.; Deretic, V.; Feng, P.; Akazawa, C.; et al. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat. Cell Biol. 2008, 10, 776–787. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Coppola, D.; Matsushita, N.; Cualing, H.D.; Sun, M.; Sato, Y.; Liang, C.; Jung, J.U.; Cheng, J.Q.; Mulé, J.J.; et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat. Cell Biol. 2007, 9, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Etxebarria, A.; Terrones, O.; Yamaguchi, H.; Landajuela, A.; Landeta, O.; Antonsson, B.; Wang, H.G.; Basañez, G. Endophilin B1/Bif-1 stimulates BAX activation independently from its capacity to produce large scale membrane morphological rearrangements. J. Biol. Chem. 2009, 284, 4200–4212. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Meyerkord, C.L.; Wang, H.G. Bif-1/endophilin B1: A candidate for crescent driving force in autophagy. Cell Death Differ. 2009, 16, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Cheung, A.Y.; Fu, W.Y.; Wu, C.; Zhang, M.; Mobley, W.C.; Cheung, Z.H.; Ip, N.Y. Endophilin B1 as a novel regulator of nerve growth factor/ TrkA trafficking and neurite outgrowth. J. Neurosci. 2008, 28, 9002–9012. [Google Scholar] [CrossRef] [PubMed]
- Runkle, K.B.; Meyerkord, C.L.; Desai, N.V.; Takahashi, Y.; Wang, H.G. Bif-1 suppresses breast cancer cell migration by promoting EGFR endocytic degradation. Cancer Biol. Ther. 2012, 13, 956–966. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Zhang, J.; Fan, W.; Wong, K.N.; Ding, X.; Chen, S.; Zhong, Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J. Biol. Chem. 2011, 286, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Li, X.; Zhang, P.; Chen, W.D.; Zhang, H.L.; Li, D.D.; Deng, R.; Qian, X.J.; Jiao, L.; Ji, J.; et al. Acetylation of Beclin 1 inhibits autophagosome maturation and promotes tumour growth. Nat. Commun. 2015, 6, 7215. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Le Toumelin, G.; Criollo, A.; Rain, J.C.; Gautier, F.; Juin, P.; Tasdemir, E.; Pierron, G.; Troulinaki, K.; Tavernarakis, N.; et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007, 26, 2527–2539. [Google Scholar] [CrossRef] [PubMed]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef] [PubMed]
- Davids, M.S.; Letai, A. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J. Clin. Oncol. 2012, 30, 3127–3135. [Google Scholar] [CrossRef] [PubMed]
- Oberstein, A.; Jeffrey, P.D.; Shi, Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J. Biol. Chem. 2007, 282, 13123–13132. [Google Scholar] [CrossRef] [PubMed]
- Adi-Harel, S.; Erlich, S.; Schmukler, E.; Cohen-Kedar, S.; Segev, O.; Mizrachy, L.; Hirsch, J.A.; Pinkas-Kramarski, R. Beclin 1 self-association is independent of autophagy induction by amino acid deprivation and rapamycin treatment. J. Cell. Biochem. 2010, 110, 1262–1271. [Google Scholar] [CrossRef] [PubMed]
- Ku, B.; Woo, J.S.; Liang, C.; Lee, K.H.; Jung, J.U.; Oh, B.H. An insight into the mechanistic role of Beclin 1 and its inhibition by prosurvival Bcl-2 family proteins. Autophagy 2008, 4, 519–520. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, L.; Che, K.H.; Funderburk, S.F.; Pan, L.; Pan, N.; Zhang, M.; Yue, Z.; Zhao, Y. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012, 3, 662. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Dong, J.M.; Manser, E.; Song, H. Bcl-xL and UVRAG cause a monomer-dimer switch in Beclin1. J. Biol. Chem. 2008, 283, 26274–26282. [Google Scholar] [CrossRef] [PubMed]
- Mandell, M.A.; Jain, A.; Arko-Mensah, J.; Chauhan, S.; Kimura, T.; Dinkins, C.; Silvestri, G.; Münch, J.; Kirchhoff, F.; Simonsen, A.; et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev. Cell 2014, 30, 394–409. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.; Merlot, A.M.; Krishan, S.; Jansson, P.J.; Richardson, D.R. Gene of the month: BECN1. J. Clin. Pathol. 2014, 67, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Glover, K.; Su, M.; Sinha, S.C. Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond. Protein Sci. 2016, 10, 1767–1785. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Du, Y.; Zhao, Z.; Shi, L.; Sun, L.; Huang, G.; Ye, B.; Li, C.; Dai, Z.; et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013, 32, 2685–2696. [Google Scholar] [CrossRef] [PubMed]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.; Xiong, Y. CRL4s: The CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 2009, 34, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Arias, E.E.; Chen, J.; Harper, J.W.; Walter, J.C. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 2006, 23, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Fischer, E.S.; Scrima, A.; Böhm, K.; Matsumoto, S.; Lingaraju, G.M.; Faty, M.; Yasuda, T.; Cavadini, S.; Wakasugi, M.; Hanaoka, F.; et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 2011, 147, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; O’Rourke, K.M.; Zhang, Z.; Dornan, D.; Arnott, D.; Deshaies, R.J.; Dixit, V.M. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 2004, 303, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Linardopoulou, E.V.; Parghi, S.S.; Friedman, C.; Osborn, G.E.; Parkhurst, S.M.; Trask, B.J. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007, 3, e237. [Google Scholar] [CrossRef] [PubMed]
- Bear, J.E. Sorting out endosomes in the WASH. Dev. Cell 2009, 17, 583–584. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Huang, G.; Du, Y.; Zhu, P.; Li, M.; Fan, Z. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014, 24, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Morel, E.; Dupont, N.; Codogno, P. Autophagy regulation: RNF2 targets AMBRA1. Cell Res. 2014, 24, 1029–1030. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, M.; Albiero, F.; Nazio, F.; Vescovo, T.; Perdomo, A.B.; Corazzari, M.; Marsella, C.; Piselli, P.; Gretzmeier, C.; Dengjel, J.; et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev. Cell 2014, 31, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Antonioli, M.; Albiero, F.; Piacentini, M.; Fimia, G.M. Temporal regulation of autophagy response by the CULLIN 4-AMBRA1-CULLIN 5 axis. Mol. Cell Oncol. 2015, 3, e1008304. [Google Scholar] [CrossRef] [PubMed]
- McEwan, D.G.; Dikic, I. Cullins keep autophagy under control. Dev. Cell 2014, 31, 675–676. [Google Scholar] [CrossRef] [PubMed]
- Sanjuan, M.A.; Dillon, C.P.; Tait, S.W.; Moshiach, S.; Dorsey, F.; Connell, S.; Komatsu, M.; Tanaka, K.; Cleveland, J.L.; Withoff, S.; et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 2007, 450, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Waltz, P.; Carchman, E.H.; Young, A.C.; Rao, J.; Rosengart, M.R.; Kaczorowski, D.; Zuckerbraun, B.S. Lipopolysaccaride induces autophagic signaling in macrophages via a TLR4, heme oxygenase-1 dependent pathway. Autophagy 2011, 7, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.S.; Kehrl, J.H. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J. Biol. Chem. 2008, 238, 33175–33182. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.S.; Kehrl, J.H. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci. Signal. 2010, 3, ra42. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.S.; Kehrl, J.H. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy 2010, 6, 986–987. [Google Scholar] [CrossRef] [PubMed]
- Zalckvar, E.; Berissi, H.; Mizrachy, L.; Idelchuk, Y.; Koren, I.; Eisenstein, M.; Sabanay, H.; Pinkas-Kramarski, R.; Kimchi, A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009, 10, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Lopez, G.Y.; Kim, C.K.; Alvarez, A.; Duncan, C.G.; Nishikawa, R.; Nagane, M.; Su, A.J.; Auron, P.E.; Hedberg, M.L.; et al. EGFR phosphorylation of DCBLD2 recruits TRAF6 and stimulates AKT-promoted tumorigenesis. J. Clin. Investig. 2014, 124, 3741–3756. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Pattingre, S.; Sinha, S.; Bassik, M.; Levine, B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol. Cell 2008, 30, 678–688. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Deng, L.; Hong, M.; Akkaraju, G.R.; Inoue, J.; Chen, Z.J. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 2001, 412, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 2012, 246, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Young, A.R.; Chan, E.Y.; Hu, X.W.; Köchl, R.; Crawshaw, S.G.; High, S.; Hailey, D.W.; Lippincott-Schwartz, J.; Tooze, S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006, 119, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.W.; Liao, H.M.; Peng, W.H.; Lin, H.R.; Chen, C.H.; Chen, G.C. Atg9 interacts with dTRAF2/TRAF6 to regulate oxidative stress-induced JNK activation and autophagy induction. Dev. Cell 2013, 27, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Strappazzon, F.; Antonioli, M.; Bielli, P.; Cianfanelli, V.; Bordi, M.; Gretzmeier, C.; Dengjel, J.; Piacentini, M.; Fimia, G.M.; et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat. Cell Biol. 2013, 15, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [PubMed]
- De Poot, S.A.H.; Tian, G.; Finley, D. Meddling with fate: The proteasomal deubiquitinating enzymes. J. Mol. Biol. 2017, 429, 3525–3545. [Google Scholar] [CrossRef] [PubMed]
- Vaden, J.H.; Bhattacharyya, B.J.; Chen, P.C.; Watson, J.A.; Marshall, A.G.; Phillips, S.E.; Wilson, J.A.; King, G.D.; Miller, R.J.; Wilson, S.M. Ubiquitin-specific protease 14 regulates c-Jun N-terminal kinase signaling at the neuromuscular junction. Mol. Neurodegener. 2015, 10, 3. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shan, B.; Lee, B.H.; Zhu, K.; Zhang, T.; Sun, H.; Liu, M.; Shi, L.; Liang, W.; Qian, L.; et al. Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. eLife 2015, 4, e10510. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Shan, B.; Sun, H.; Xiao, J.; Zhu, K.; Xie, X.; Li, X.; Liang, W.; Lu, X.; Qian, L.; et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 2016, 30, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Vogel, R.I.; Pulver, T.; Heilmann, W.; Mooneyham, A.; Mullany, S.; Zhao, X.; Shahi, M.; Richter, J.; Klein, M.; Chen, L.; et al. USP14 is a predictor of recurrence in endometrial cancer and a molecular target for endometrial cancer treatment. Oncotarget 2016, 7, 30962–30976. [Google Scholar] [CrossRef] [PubMed]
- Fajner, V.; Maspero, E.; Polo, S. Targeting HECT-type E3 ligases—Insights from catalysis, regulation and inhibitors. FEBS Lett. 2017, 591, 2636–2647. [Google Scholar] [CrossRef] [PubMed]
- Platta, H.W.; Abrahamsen, H.; Thoresen, S.B.; Stenmark, H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem. J. 2012, 441, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Aita, V.M.; Liang, X.H.; Murty, V.V.; Pincus, D.L.; Yu, W.; Cayanis, E.; Kalachikov, S.; Gilliam, T.C.; Levine, B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999, 59, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, Y.; Lu, C.; Zhang, L. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expression. Pathol. Oncol. Res. 2009, 15, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Pitiakoudis, M.; Gatter, K.C.; Harris, A.L. Beclin 1 over- and underexpression in colorectal cancer: Distinct patterns relate to prognosis and tumour hypoxia. Br. J. Cancer 2010, 103, 1209–1214. [Google Scholar] [CrossRef] [PubMed]
- Behrends, C.; Sowa, M.E.; Gygi, S.P.; Harper, J.W. Network organization of the human autophagy system. Nature 2010, 466, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Wei, J.; Childress, C.; Shaw, J.H.t.; Peng, K.; Shao, G.; Yang, W.; Lin, Q. The E3 ubiquitin ligase NEDD4 is an LC3-interactive protein and regulates autophagy. Autophagy 2017, 13, 522–537. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Zheng, Y.; Wu, K.P.; Schulman, B.A. Insights into links between autophagy and the ubiquitin system from the structure of LC3B bound to the LIR motif from the E3 ligase NEDD4. Protein Sci. 2017, 26, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Zhu, N.; Chen, S.; Zhao, P.; Ren, H.; Zhu, S.; Tang, H.; Zhu, Y.; Qi, Z. E3 Ubiquitin Ligase Nedd4 Promotes Japanese Encephalitis Virus Replication by Suppressing Autophagy in Human Neuroblastoma Cells. Sci. Rep. 2017, 7, 45375. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fan, C.D.; Wang, X. Regulation of Mdm2 protein stability and the p53 response by NEDD4-1 E3 ligase. Oncogene 2015, 34, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Yang, Y.; Lu, M.; Xu, L.; Liu, F.; Yuan, Z.; Bao, Q.; Jiang, Z.; Xu, X.; Guo, X.; et al. Escape, or Vanish: Control the Fate of p53 through MDM2-Mediated Ubiquitination. Anticancer Agents Med. Chem. 2015, 16, 174–189. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.; Ash, D.; Shaha, C. Beclin-1-p53 interaction is crucial for cell fate determination in embryonal carcinoma cells. J. Cell. Mol. Med. 2014, 18, 2275–2286. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, K.; Pathak, C. Expression of cFLIPL Determines the Basal Interaction of Bcl-2 With Beclin-1 and Regulates p53 Dependent Ubiquitination of Beclin-1 During Autophagic Stress. J. Cell. Biochem. 2016, 117, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Pu, J.L.; Krafft, P.R.; Zhang, J.M.; Chen, S. The Molecular Mechanisms Between Autophagy and Apoptosis: Potential Role in Central Nervous System Disorders. Cell. Mol. Neurobiol. 2015, 35, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Niso-Santano, M.; Baehrecke, E.H.; Kroemer, G. Self-consumption: The interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014, 15, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Hassink, G.C.; Zhao, B.; Sompallae, R.; Altun, M.; Gastaldello, S.; Zinin, N.V.; Masucci, M.G.; Lindsten, K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009, 10, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Hahn, A.A.; Hu, S.; Yang, X. The USP19 deubiquitinase regulates the stability of c-IAP1 and c-IAP2. J. Biol. Chem. 2011, 286, 35380–35387. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R.F. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Jin, S.; Wang, R.F. The BECN1-USP19 axis plays a role in the crosstalk between autophagy and antiviral immune responses. Autophagy 2016, 12, 1210–1211. [Google Scholar] [CrossRef] [PubMed]
- Rohatgi, R.A.; Shaw, L.M. An autophagy-independent function for Beclin 1 in cancer. Mol. Cell. Oncol. 2016, 3, e1030539. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Xia, H.; Kim, M.; Xu, L.; Li, Y.; Zhang, L.; Cai, Y.; Norberg, H.V.; Zhang, T.; Furuya, T.; et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011, 147, 223–234. [Google Scholar] [CrossRef] [PubMed]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Thoresen, S.B.; Pedersen, N.M.; Liestøl, K.; Stenmark, H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp. Cell Res. 2010, 316, 3368–3378. [Google Scholar] [CrossRef] [PubMed]
- Sowa, M.E.; Bennett, E.J.; Gygi, S.P.; Harper, J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009, 138, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Nazarko, V.Y.; Zhong, Q. ULK1 targets Beclin-1 in autophagy. Nat. Cell Biol. 2013, 15, 727–728. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Luo, K.; Zhang, L.; Cheville, J.C.; Lou, Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell 2010, 140, 384–396. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Li, S.; Qin, Y.; Wang, X.; Yang, Y.; Bai, H.; Zhou, L.; Zhao, C.; Wang, C. Spautin-1, a novel autophagy inhibitor, enhances imatinib-induced apoptosis in chronic myeloid leukemia. Int. J. Oncol. 2014, 44, 1661–1668. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.J.; Valdes, Y.R.; Peart, T.M.; Fazio, E.N.; Bertrand, M.; McGee, J.; Préfontaine, M.; Sugimoto, A.; DiMattia, G.E.; Shepherd, T.G. Combination of AKT inhibition with autophagy blockade effectively reduces ascites-derived ovarian cancer cell viability. Carcinogenesis 2014, 35, 1951–1961. [Google Scholar] [CrossRef] [PubMed]
- Into, T.; Inomata, M.; Takayama, E.; Takigawa, T. Autophagy in regulation of Toll-like receptor signaling. Cell Signal. 2012, 24, 1150–1162. [Google Scholar] [CrossRef] [PubMed]
- Dove, K.K.; Klevit, R.E. RING-Between-RING E3 Ligases: Emerging Themes amid the Variations. J. Mol. Biol. 2017, 429, 3363–3375. [Google Scholar] [CrossRef] [PubMed]
- Chuang, T.H.; Ulevitch, R.J. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat. Immunol. 2004, 5, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Fearns, C.; Pan, Q.; Mathison, J.C.; Chuang, T.H. Triad3A regulates ubiquitination and proteasomal degradation of RIP1 following disruption of Hsp90 binding. J. Biol. Chem. 2006, 281, 34592–34600. [Google Scholar] [CrossRef] [PubMed]
- Nakhaei, P.; Mesplede, T.; Solis, M.; Sun, Q.; Zhao, T.; Yang, L.; Chuang, T.H.; Ware, C.F.; Lin, R.; Hiscott, J. The E3 ubiquitin ligase Triad3A negatively regulates the RIG-I/MAVS signaling pathway by targeting TRAF3 for degradation. PLoS Pathog. 2009, 5, e1000650. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Feng, K.; Zhao, X.; Huang, S.; Cheng, Y.; Qian, L.; Wang, Y.; Sun, H.; Jin, M.; Chuang, T.H.; et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy 2014, 10, 2239–2250. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, Y.; Qian, L.; Wang, X.; Gu, H.; Dong, X.; Huang, S.; Jin, M.; Ge, H.; Xu, C.; et al. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget 2016, 7, 51174–51183. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Bath, M.L.; Turner, M.L.; Vandenberg, C.J.; Bouillet, P.; Metcalf, D.; Scott, C.L.; Cory, S. Elevated Mcl-1 perturbs lymphopoiesis, promotes transformation of hematopoietic stem/progenitor cells, and enhances drug resistance. Blood 2010, 116, 3197–3207. [Google Scholar] [CrossRef] [PubMed]
- Aichberger, K.J.; Mayerhofer, M.; Krauth, M.T.; Skvara, H.; Florian, S.; Sonneck, K.; Akgul, C.; Derdak, S.; Pickl, W.F.; Wacheck, V.; et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): Evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood 2005, 105, 3303–3311. [Google Scholar] [CrossRef] [PubMed]
- Cho-Vega, J.H.; Rassidakis, G.Z.; Admirand, J.H.; Oyarzo, M.; Ramalingam, P.; Paraguya, A.; McDonnell, T.J.; Amin, H.M.; Medeiros, L.J. MCL-1 expression in B-cell non-Hodgkin’s lymphomas. Hum. Pathol. 2004, 35, 1095–1100. [Google Scholar] [CrossRef] [PubMed]
- Perciavalle, R.M.; Stewart, D.P.; Koss, B.; Lynch, J.; Milasta, S.; Bathina, M.; Temirov, J.; Cleland, M.M.; Pelletier, S.; Schuetz, J.D.; et al. Anti-apoptotic MCL-1 localizes to the mitochondrial matrix and couples mitochondrial fusion to respiration. Nat. Cell Biol. 2012, 14, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Elgendy, M.; Sheridan, C.; Brumatti, G.; Martin, S.J. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol. Cell 2011, 42, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Elgendy, M.; Ciro, M.; Abdel-Aziz, A.K.; Belmonte, G.; Dal Zuffo, R.; Mercurio, C.; Miracco, C.; Lanfrancone, L.; Foiani, M.; Minucci, S. Beclin 1 restrains tumorigenesis through Mcl-1 destabilization in an autophagy-independent reciprocal manner. Nat. Commun. 2014, 5, 5637. [Google Scholar] [CrossRef] [PubMed]
- Meyerovich, K.; Violato, N.M.; Fukaya, M.; Dirix, V.; Pachera, N.; Marselli, L.; Marchetti, P.; Strasser, A.; Eizirik, D.L.; Cardozo, A.K. MCL-1 Is a Key Antiapoptotic Protein in Human and Rodent Pancreatic β-Cells. Diabetes 2017, 66, 2446–2458. [Google Scholar] [CrossRef] [PubMed]
- Trivigno, D.; Essmann, F.; Huber, S.M.; Rudner, J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia 2012, 14, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Mojsa, B.; Lassot, I.; Desagher, S. Mcl-1 ubiquitination: Unique regulation of an essential survival protein. Cells 2014, 3, 418–437. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Q.; Gao, W.; Du, F.; Wang, X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell 2005, 21, 71085–71095. [Google Scholar] [CrossRef] [PubMed]
- Elgendy, M.; Minucci, S. A novel autophagy-independent, oncosuppressive function of BECN1: Degradation of MCL1. Autophagy 2015, 11, 581–582. [Google Scholar] [CrossRef] [PubMed]
- Murtaza, M.; Jolly, L.A.; Gecz, J.; Wood, S.A. La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 2015, 72, 2075–2089. [Google Scholar] [CrossRef] [PubMed]
- Eckl, J.M.; Richter, K. Functions of the Hsp90 chaperone system: Lifting client proteins to new heights. Int. J. Biochem. Mol. Biol. 2013, 4, 157–165. [Google Scholar] [PubMed]
- Miyata, Y. Hsp90 inhibitor geldanamycin and its derivatives as novel cancer chemotherapeutic agents. Curr. Pharm. Des. 2005, 11, 1131–1138. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Liu, J.; Hsu, L.C.; Luo, Y.; Xiang, R.; Chuang, T.H. Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J. 2011, 25, 2700–2710. [Google Scholar] [CrossRef] [PubMed]
- Ehrlich, E.S.; Wang, T.; Luo, K.; Xiao, Z.; Niewiadomska, A.M.; Martinez, T.; Xu, W.; Neckers, L.; Yu, X.F. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2009, 106, 20330–20335. [Google Scholar] [CrossRef] [PubMed]
- Samant, R.S.; Clarke, P.A.; Workman, P. E3 ubiquitin ligase Cullin-5 modulates multiple molecular and cellular responses to heat shock protein 90 inhibition in human cancer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 6834–6839. [Google Scholar] [CrossRef] [PubMed]
- Kochhar, A.; Kopelovich, L.; Sue, E.; Guttenplan, J.B.; Herbert, B.S.; Dannenberg, A.J.; Subbaramaiah, K. p53 modulates Hsp90 ATPase activity and regulates aryl hydrocarbon receptor signaling. Cancer Prev. Res. (Phila) 2014, 7, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Okayama, S.; Kopelovich, L.; Balmus, G.; Weiss, R.S.; Herbert, B.S.; Dannenberg, A.J.; Subbaramaiah, K. p53 protein regulates Hsp90 ATPase activity and thereby Wnt signaling by modulating Aha1 expression. J. Biol. Chem. 2014, 289, 6513–6525. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Uesugi, M.; Takase, K.; Miyamoto, N.; Sawada, K. Hsp90 inhibitor geldanamycin attenuates the cytotoxicity of sunitinib in cardiomyocytes via inhibition of the autophagy pathway. Toxicol. Appl. Pharmacol. 2017, 329, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.H.; Dorsey, F.C.; Joshi, A.; Hennessy-Walters, K.M.; Rose, K.L.; McCastlain, K.; Zhang, J.; Iyengar, R.; Jung, C.H.; Suen, D.F.; et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol. Cell 2011, 43, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Carbonero, R.; Carnero, A.; Paz-Ares, L. Inhibition of HSP90 molecular chaperones: Moving into the clinic. Lancet Oncol. 2013, 14, e358–e369. [Google Scholar] [CrossRef]
- Kim, T.; Keum, G.; Pae, A.N. Discovery and development of heat shock protein 90 inhibitors as anticancer agents: A review of patented potent geldanamycin derivatives. Expert Opin. Ther. Pat. 2013, 23, 919–943. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Chen, Z.; Yu, F.; Chen, Q.; Tian, Y.; Ma, S.; Wang, T.; Liu, X. Hsp90 regulates autophagy and plays a role in cancer therapy. Tumour Biol. 2016, 37, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Liang, C. Negative regulation of autophagy. Cell Death Differ. 2010, 17, 1807–1815. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Chen, R.H. KLHL20 links the ubiquitin-proteasome system to autophagy termination. Autophagy 2016, 12, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.R.; Yuan, W.C.; Ho, H.C.; Chen, C.H.; Shih, H.M.; Chen, R.H. The Cullin 3 substrate adaptor KLHL20 mediates DAPK ubiquitination to control interferon responses. EMBO J. 2010, 29, 1748–1761. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Lin, Y.C.; Chen, Y.H.; Chen, C.M.; Pang, L.Y.; Chen, H.A.; Wu, P.R.; Lin, M.Y.; Jiang, S.T.; Tsai, T.F.; et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Nazio, F.; Carinci, M.; Valacca, C.; Bielli, P.; Strappazzon, F.; Antonioli, M.; Ciccosanti, F.; Rodolfo, C.; Campello, S.; Fimia, G.M.; et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J. Cell Biol. 2016, 215, 841–856. [Google Scholar] [CrossRef] [PubMed]
- Neel, B.A.; Lin, Y.; Pessin, J.E. Skeletal muscle autophagy: A new metabolic regulator. Trends Endocrinol. Metab. 2013, 12, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Sandri, M.; Coletto, L.; Grumati, P.; Bonaldo, P. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J. Cell Sci. 2013, 126, 5325–5333. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Klionsky, D.J. Downregulation of autophagy through CUL3-KLHL20-mediated turnover of the ULK1 and PIK3C3/VPS34 complexes. Autophagy 2016, 12, 1071–1072. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; Liu, C.C.; Chen, R.H. Cul3-KLHL20 ubiquitin ligase: Physiological functions, stress responses, and disease implications. Cell Div. 2016, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fiskus, W.; Yong, B.; Atadja, P.; Takahashi, Y.; Pandita, T.K.; Wang, H.G.; Bhalla, K.N. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 6841–6846. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Zhang, T.; Xiao, J.; Zhu, K.; Wei, R.; Wu, Z.; Meng, H.; Li, Y.; Yuan, J. Modification of BECN1 by ISG15 plays a crucial role in autophagy regulation by type I IFN/interferon. Autophagy 2015, 11, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.J.; Chen, F.Z.; Chen, H.C.; Wan, X.X.; Zhou, X.; Fang, Q.; Zhang, D.Z. ISG15 inhibits cancer cell growth and promotes apoptosis. Int. J. Mol. Med. 2017, 39, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Hermann, M.; Bogunovic, D. ISG15: In Sickness and in Health. Trends Immunol. 2017, 38, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Durfee, L.A.; Huibregtse, J.M. The ISG15 conjugation system. Methods Mol. Biol. 2012, 832, 141–149. [Google Scholar] [PubMed]
- Basters, A.; Geurink, P.P.; Röcker, A.; Witting, K.F.; Tadayon, R.; Hess, S.; Semrau, M.S.; Storici, P.; Ovaa, H.; Knobeloch, K.P.; et al. Structural basis of the specificity of USP18 toward ISG15. Nat. Struct. Mol. Biol. 2017, 24, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Basters, A.; Geurink, P.P.; El Oualid, F.; Ketscher, L.; Casutt, M.S.; Krause, E.; Ovaa, H.; Knobeloch, K.P.; Fritz, G. Molecular characterization of ubiquitin-specific protease 18 reveals substrate specificity for interferon-stimulated gene 15. FEBS J. 2014, 281, 1918–1928. [Google Scholar] [CrossRef] [PubMed]
- Jordan, T.X.; Randall, G. Manipulation or capitulation: Virus interactions with autophagy. Microbes Infect. 2012, 14, 126–139. [Google Scholar] [CrossRef] [PubMed]
- Rey-Jurado, E.; Riedel, C.A.; González, P.A.; Bueno, S.M.; Kalergis, A.M. Contribution of autophagy to antiviral immunity. FEBS Lett. 2015, 589, 3461–3470. [Google Scholar] [CrossRef] [PubMed]
- Falvey, C.M.; O’Donovan, T.R.; El-Mashed, S.; Nyhan, M.J.; O’Reilly, S.; McKenna, S.L. UBE2L6/UBCH8 and ISG15 attenuate autophagy in esophageal cancer cells. Oncotarget 2017, 8, 23479–23491. [Google Scholar] [CrossRef] [PubMed]
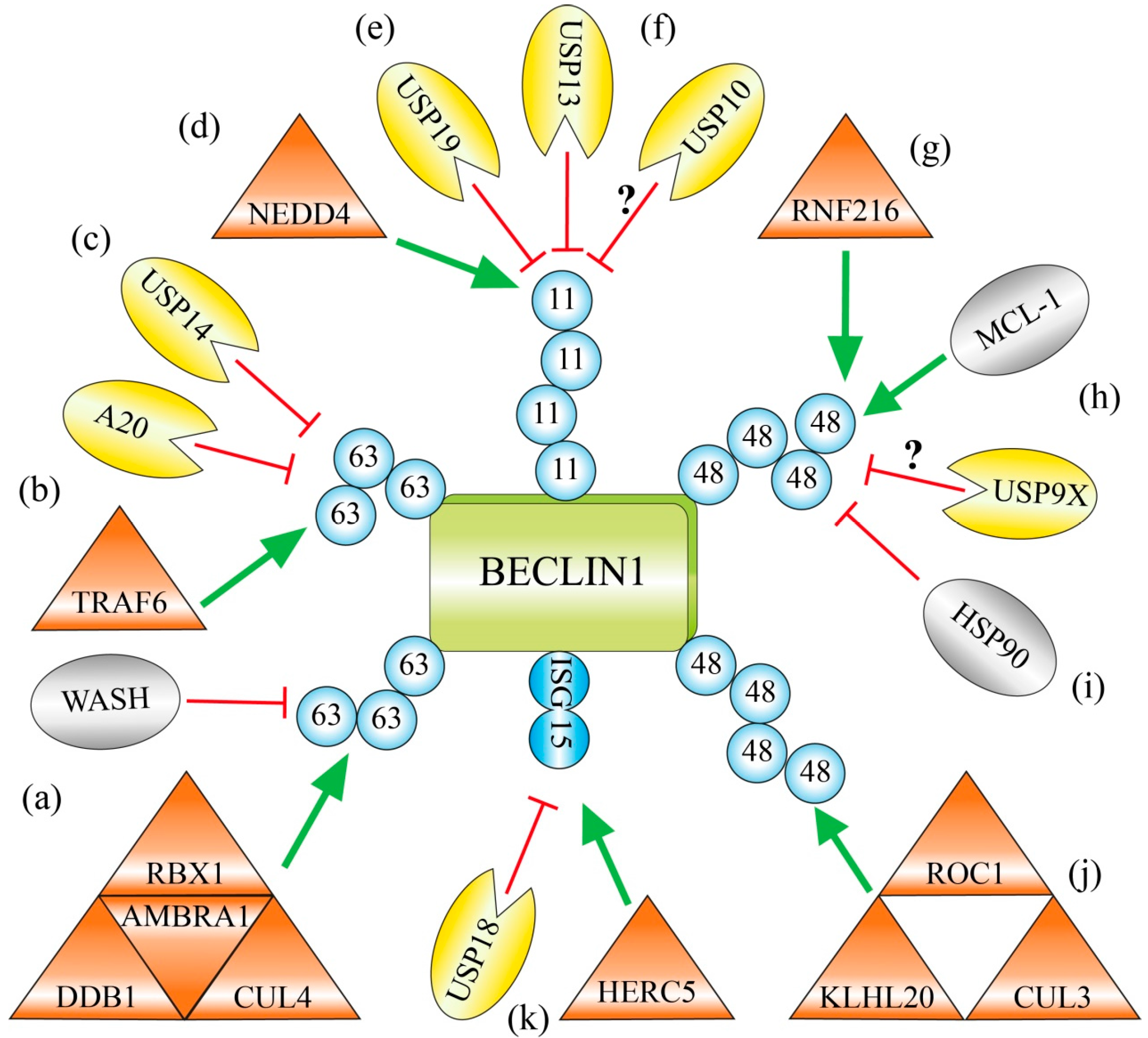
| Name | Type | Target Lys in BECLIN 1 | Modification of BECLIN 1 | Effect on Modification | Effect on BECLIN 1 | Effect on Autophagy |
|---|---|---|---|---|---|---|
| CUL4-complex | E3 (CUL) | Lys437 | PolyUb (Lys63) | Synthesis | Strong VPS34 binding | + |
| WASH | WASP | Lys437 | PolyUb (Lys63) | Inhibition | Weak VPS34 binding | − |
| TRAF6 | E3 (RING) | Lys117 | PolyUb (Lys63) | Synthesis | no BCL-2 binding | + |
| A20 | DUB | Lys117 | PolyUb (Lys63) | Hydrolysis | BCL-2 binding | − |
| USP14 | DUB | [?] | PolyUb (Lys63) | Hydrolysis | weak ATG14L/UVRAG binding | + |
| NEDD4 | E3 (HECT) | [?] | PolyUb (Lys11) | Synthesis | Degradation | − [?] |
| USP19 | DUB | Lys437 | PolyUb (Lys11) | Hydrolysis | Stabilisation | + |
| USP13 | DUB | [?] | PolyUb (Lys11) | Hydrolysis | Stabilisation | + |
| USP10 | DUB | [?] | PolyUb (Lys11) [?] | Hydrolysis | Stabilisation | + |
| RNF216 | E3 (RBR) | [?] | PolyUb (Lys48) | Synthesis | Degradation | − |
| MCL-1 | BCL-2 Family | [?] | PolyUb (Lys[?]) | Induction | Degradation | − |
| USP9X | DUB | [?] | PolyUb (Lys[?]) | Hydrolysis | Stabilisation | + |
| HSP90 | HSP | [?] | PolyUb (Lys48) | Inhibition | Stabilisation | + |
| CUL3-complex | E3 (CUL) | [?] | PolyUb (Lys48) | Synthesis | Degradation | − |
| HERC5 | E3 (for ISG) | Lys117/263/265/266 | ISGylation | Synthesis | Weak VPS34 binding [?] | − |
| PolyUb (Lys63) | Inhibition | |||||
| USP18 | DUB (for ISG) | Lys117/263/265/266 | ISGylation | Hydrolysis | Strong VPS34 binding [?] | + |
| PolyUb (Lys63) | no Inhibition |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boutouja, F.; Brinkmeier, R.; Mastalski, T.; El Magraoui, F.; Platta, H.W. Regulation of the Tumor-Suppressor BECLIN 1 by Distinct Ubiquitination Cascades. Int. J. Mol. Sci. 2017, 18, 2541. https://doi.org/10.3390/ijms18122541
Boutouja F, Brinkmeier R, Mastalski T, El Magraoui F, Platta HW. Regulation of the Tumor-Suppressor BECLIN 1 by Distinct Ubiquitination Cascades. International Journal of Molecular Sciences. 2017; 18(12):2541. https://doi.org/10.3390/ijms18122541
Chicago/Turabian StyleBoutouja, Fahd, Rebecca Brinkmeier, Thomas Mastalski, Fouzi El Magraoui, and Harald W. Platta. 2017. "Regulation of the Tumor-Suppressor BECLIN 1 by Distinct Ubiquitination Cascades" International Journal of Molecular Sciences 18, no. 12: 2541. https://doi.org/10.3390/ijms18122541




