Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD)
Abstract
:1. Introduction
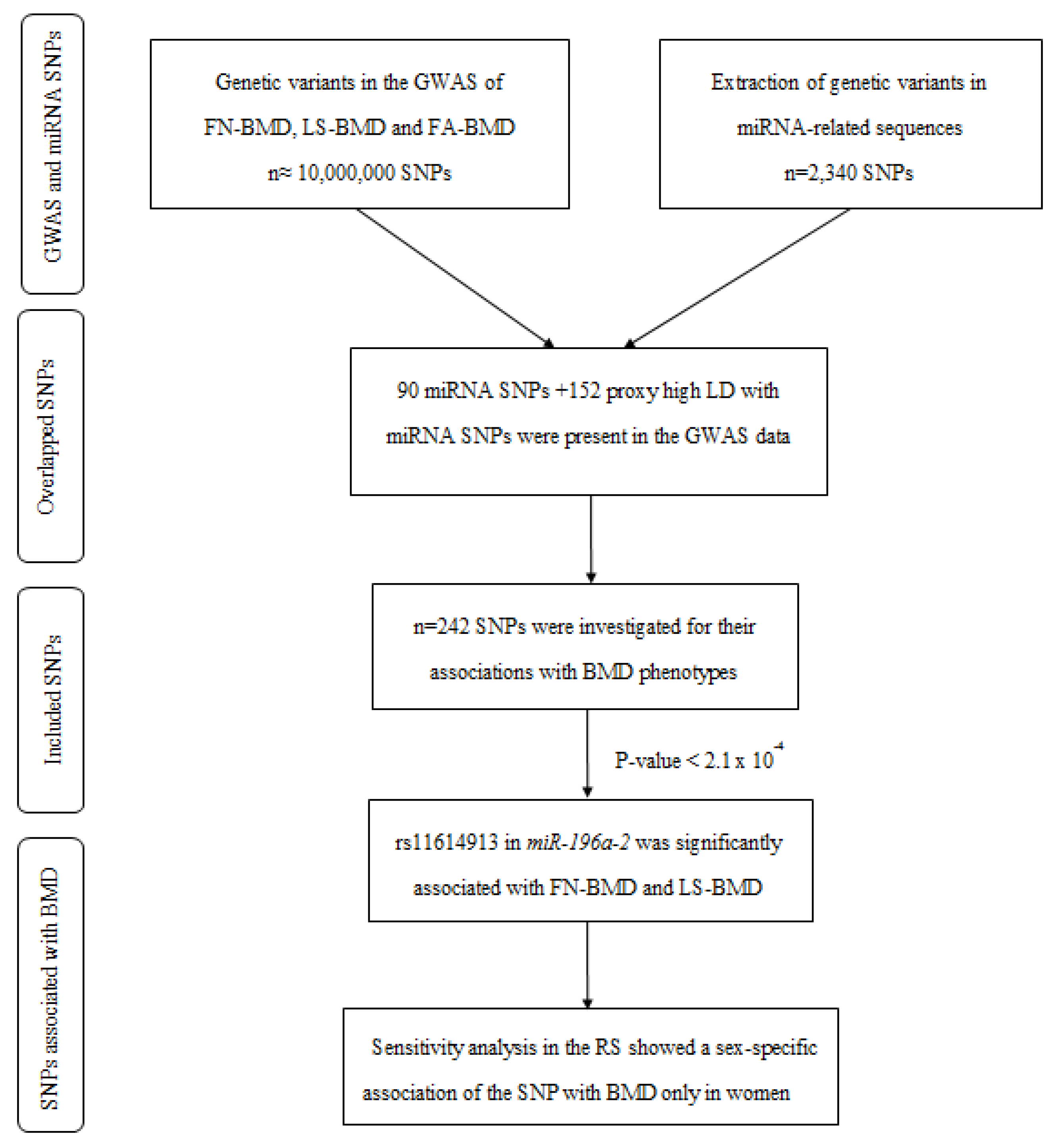
2. Results
2.1. A Variant in miR-196a-2 Associates with BMD
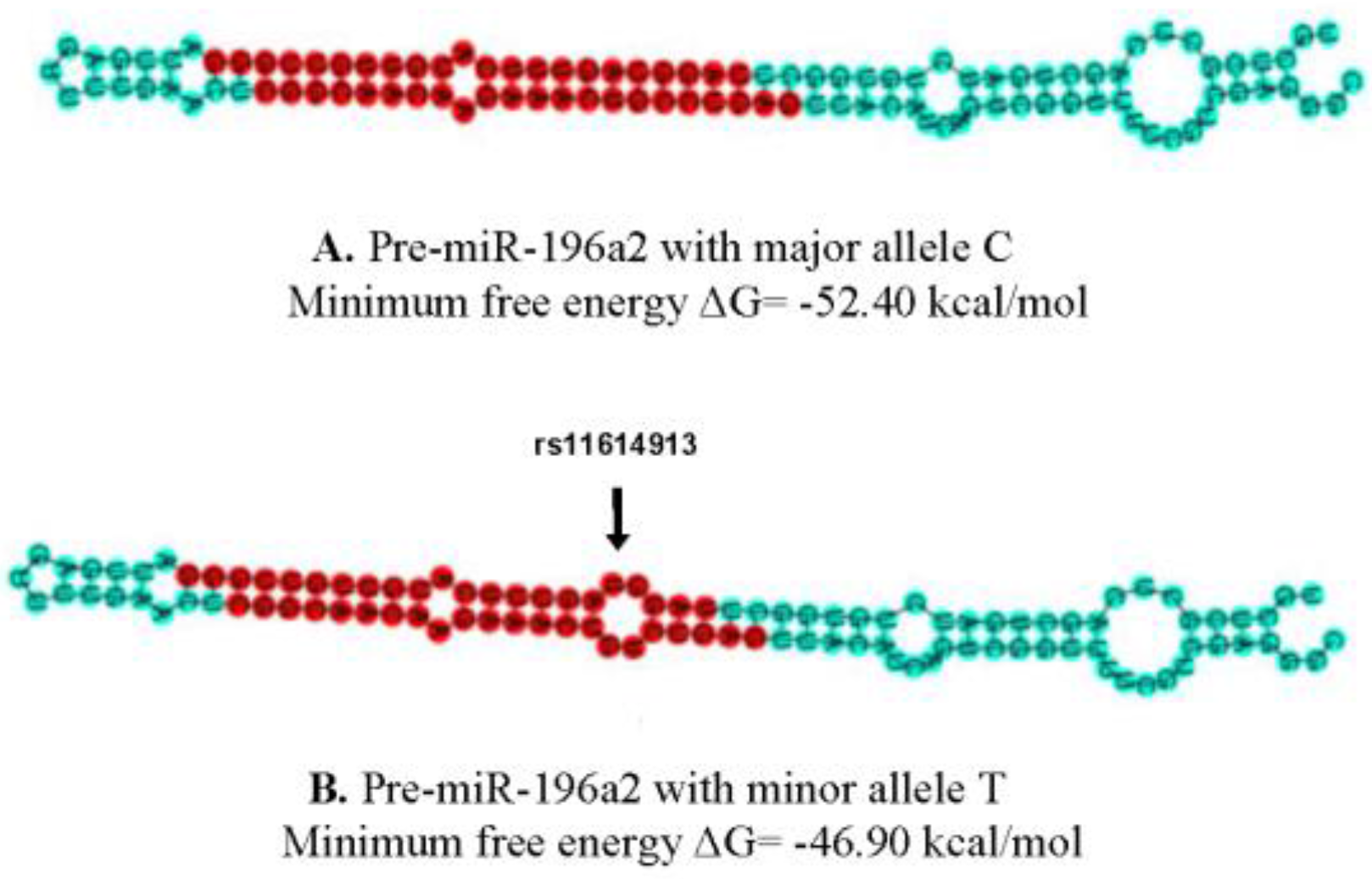
2.2. The Potential Impact of rs11614913 on the miR-196a-2 Structure and Function
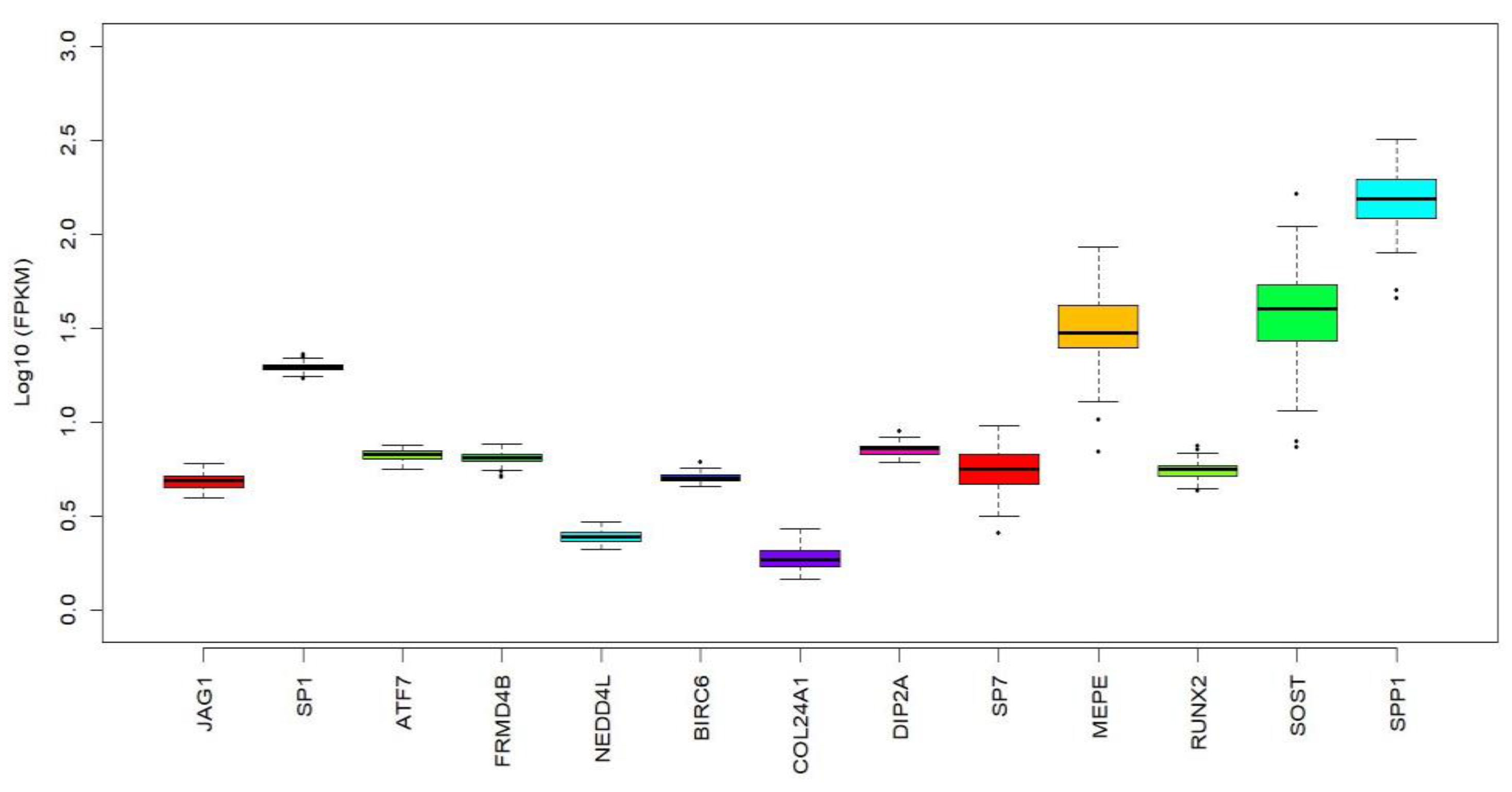
2.3. Associaton of miR-196a-2 Target Genes with BMD
2.4. Sensitivity Analyses for rs11614913 in miR-196a-2 Using the Rotterdam Study Data
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Association Studies on BMD Phenotypes
4.2. Identification of Genetic Variants in miRNA-Encoding Sequences
4.3. miRNA Target Genes Associated with BMD Phenotypes
4.4. The Variant Effect on the Pre-miRNA Structure
4.5. The Rotterdam Study Data
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association studies |
| GEFOS | Genetic factors for osteoporosis |
| BMD SNP | Bone mineral density Single nucleotide polymorphism |
| FN-BMD | Femoral neck bone mineral density |
| LS-BMD | Lumbar spine bone mineral density |
| FA-BMD | Forearm bone mineral density |
| miRNA | microRNA |
| WHR | Waist to hip ratio |
| RS | Rotterdam Study |
| DXA | Dual X-ray Absorptiometry |
| MFE | Minimum free energy |
| LD | Linkage disequilibrium |
| DHEA | Dehydroepiandrosterone |
| DHEAS | Dehydroepiandrosterone sulfate |
| IQR | Interquartile range |
References
- Hernlund, E.; Svedbom, A.; Ivergard, M.; Compston, J.; Cooper, C.; Stenmark, J.; McCloskey, E.V.; Jonsson, B.; Kanis, J.A. Osteoporosis in the European Union: Medical management, epidemiology and economic burden. A report prepared in collaboration with the International Osteoporosis Foundation (IOF) and the European Federation of Pharmaceutical Industry Associations (EFPIA). Arch Osteoporos 2013, 8, 136. [Google Scholar] [CrossRef] [PubMed]
- Rivadeneira, F.; Makitie, O. Osteoporosis and bone mass disorders: From gene pathways to treatments. Trends Endocrinol. Metab. 2016, 27, 262–281. [Google Scholar] [CrossRef] [PubMed]
- Rivadeneira, F.; Styrkarsdottir, U.; Estrada, K.; Halldorsson, B.V.; Hsu, Y.H.; Richards, J.B.; Zillikens, M.C.; Kavvoura, F.K.; Amin, N.; Aulchenko, Y.S.; et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009, 41, 1199–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kling, J.M.; Clarke, B.L.; Sandhu, N.P. Osteoporosis prevention, screening, and treatment: A review. J. Womens Health 2014, 23, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Blake, G.M.; Fogelman, I. The role of DXA bone density scans in the diagnosis and treatment of osteoporosis. Postgrad. Med. J. 2007, 83, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Stewart, T.L.; Ralston, S.H. Role of genetic factors in the pathogenesis of osteoporosis. J. Endocrinol. 2000, 166, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.F.; Forgetta, V.; Hsu, Y.H.; Estrada, K.; Rosello-Diez, A.; Leo, P.J.; Dahia, C.L.; Park-Min, K.H.; Tobias, J.H.; Kooperberg, C.; et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 2015, 526, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.L.; Guo, Y.; Zhang, Y.; Dong, S.S.; Xu, W.; Hao, R.H.; Chen, X.F.; Yan, H.; Yang, S.Y.; Yang, T.L. A functional SNP regulated by miR-196a-3p in the 3’ UTR of FGF2 is associated with bone mineral density in the Chinese population. Hum. Mutat. 2017, 38, 725–735. [Google Scholar] [CrossRef] [PubMed]
- Dole, N.S.; Kapinas, K.; Kessler, C.B.; Yee, S.P.; Adams, D.J.; Pereira, R.C.; Delany, A.M. A single nucleotide polymorphism in osteonectin 3′ untranslated region regulates bone volume and is targeted by miR-433. J. Bone Miner. Res. 2015, 30, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.J.; Liao, L.; Yang, L.; Li, Y.; Jiang, T.J. MiR-125a TNF receptor-associated factor 6 to inhibit osteoclastogenesis. Exp. Cell Res. 2014, 321, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Jemtland, R.; Holden, M.; Reppe, S.; Olstad, O.K.; Reinholt, F.P.; Gautvik, V.T.; Refvem, H.; Frigessi, A.; Houston, B.; Gautvik, K.M. Molecular disease map of bone characterizing the postmenopausal osteoporosis phenotype. J. Bone Miner. Res. 2011, 26, 1793–1801. [Google Scholar] [CrossRef] [PubMed]
- Reppe, S.; Refvem, H.; Gautvik, V.T.; Olstad, O.K.; Hovring, P.I.; Reinholt, F.P.; Holden, M.; Frigessi, A.; Jemtland, R.; Gautvik, K.M. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone 2010, 46, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Bushati, N.; Cohen, S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007, 23, 175–205. [Google Scholar] [CrossRef] [PubMed]
- Ardekani, A.M.; Naeini, M.M. The role of microRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010, 2, 161–179. [Google Scholar] [PubMed]
- Tufekci, K.U.; Oner, M.G.; Meuwissen, R.L.; Genc, S. The role of microRNAs in human diseases. Methods Mol. Biol. 2014, 1107, 33–50. [Google Scholar] [PubMed]
- Zhang, Y.; Lu, Y.J.; Yang, B.F. Potential role of microRNAs in human diseases and the exploration on design of small molecule agents. Acta Pharm. Sin. 2007, 42, 1115–1121. [Google Scholar]
- Lu, J.; Clark, A.G. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012, 22, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Dole, N.S.; Delany, A.M. microRNA variants as genetic determinants of bone mass. Bone 2016, 84, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Huang, X.; Lu, C.; Cairo, M.S.; Zhou, X. microRNA-146a and microRNA-146b regulate human dendritic cell apoptosis and cytokine production by targeting TRAF6 and IRAK1 proteins. J. Biol. Chem. 2015, 290, 2831–2841. [Google Scholar] [CrossRef] [PubMed]
- Gravallese, E.M.; Galson, D.L.; Goldring, S.R.; Auron, P.E. The role of TNF-receptor family members and other TRAF-dependent receptors in bone resorption. Arthritis Res. 2001, 3, 6–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Jin, H.M.; Kim, K.; Song, I.; Youn, B.U.; Matsuo, K.; Kim, N. The mechanism of osteoclast differentiation induced by IL-1. J. Immunol. 2009, 183, 1862–1870. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Wei, D.; Xie, B.; Ni, J.; Xuan, D.; Zhang, J. Effect and possible mechanism of network between microRNAs and RUNX2 gene on human dental follicle cells. J. Cell. Biochem. 2014, 115, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Nakasa, T.; Shibuya, H.; Nagata, Y.; Niimoto, T.; Ochi, M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheumatol. 2011, 63, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, A.; Cuvellier, S.; Naguibneva, I.; Duquet, A.; Moss, E.G.; Harel-Bellan, A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007, 21, 1125–1138. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.Q.; Gordon, J.A.; Beloti, M.M.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. A network connecting Runx2, SATB2, and the miR-23a~27a~24-2 cluster regulates the osteoblast differentiation program. Proc. Natl. Acad. Sci. USA 2010, 107, 19879–19884. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Liu, C.; Liu, W.; Wu, Y.; Ma, Z.; Chen, H.; Guo, A.Y. An update of miRNASNP database for better SNP selection by GWAS data, miRNA expression and online tools. Database (Oxford) 2015, 2015. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, R.; Bernhart, S.H.; Honer Zu Siederdissen, C.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Xu, J.; Zhao, D.; Geng, M.; Ge, H.; Fu, L.; Zhu, Z. Somatic mutation of the SNP rs11614913 and its association with increased miR-196a2 Expression in Breast Cancer. DNA Cell Biol. 2016, 35, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, J.; Tian, T.; Zhou, X.; Gu, H.; Xu, L.; Zeng, Y.; Miao, R.; Jin, G.; Ma, H.; et al. Genetic variants of miRNA sequences and non-small cell lung cancer survival. J. Clin. Investig. 2008, 118, 2600–2608. [Google Scholar] [CrossRef] [PubMed]
- Karasik, D.; Ferrari, S.L. Contribution of gender-specific genetic factors to osteoporosis risk. Ann. Hum. Genet. 2008, 72, 696–714. [Google Scholar] [CrossRef] [PubMed]
- Naganathan, V.; Macgregor, A.; Snieder, H.; Nguyen, T.; Spector, T.; Sambrook, P. Gender differences in the genetic factors responsible for variation in bone density and ultrasound. J. Bone Miner. Res. 2002, 17, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Corsi, K.A.; Pollett, J.B.; Phillippi, J.A.; Usas, A.; Li, G.; Huard, J. Osteogenic potential of postnatal skeletal muscle-derived stem cells is influenced by donor sex. J. Bone Miner. Res. 2007, 22, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Tosi, L.L.; Boyan, B.D.; Boskey, A.L. Does sex matter in musculoskeletal health? The influence of sex and gender on musculoskeletal health. J. Bone Joint Surg. Am. 2005, 87, 1631–1647. [Google Scholar] [PubMed]
- Van Wijnen, A.J.; van de Peppel, J.; van Leeuwen, J.P.; Lian, J.B.; Stein, G.S.; Westendorf, J.J.; Oursler, M.J.; Im, H.J.; Taipaleenmaki, H.; Hesse, E.; et al. MicroRNA functions in osteogenesis and dysfunctions in osteoporosis. Curr. Osteoporos Rep. 2013, 11, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gao, Y.; Cai, L.; Li, F.; Lou, Y.; Xu, N.; Kang, Y.; Yang, H. MicroRNA-221 is involved in the regulation of osteoporosis through regulates RUNX2 protein expression and osteoblast differentiation. Am. J. Transl. Res. 2017, 9, 126–135. [Google Scholar] [PubMed]
- Sun, M.; Zhou, X.; Chen, L.; Huang, S.; Leung, V.; Wu, N.; Pan, H.; Zhen, W.; Lu, W.; Peng, S. The Regulatory Roles of MicroRNAs in Bone Remodeling and Perspectives as Biomarkers in Osteoporosis. Biomed. Res. Int. 2016, 2016, 1652417. [Google Scholar] [CrossRef] [PubMed]
- De-Ugarte, L.; Yoskovitz, G.; Balcells, S.; Guerri-Fernandez, R.; Martinez-Diaz, S.; Mellibovsky, L.; Urreizti, R.; Nogues, X.; Grinberg, D.; Garcia-Giralt, N.; et al. MiRNA profiling of whole trabecular bone: Identification of osteoporosis-related changes in MiRNAs in human hip bones. BMC Med. Genom. 2015, 8, 75. [Google Scholar]
- Hackl, M.; Heilmeier, U.; Weilner, S.; Grillari, J. Circulating microRNAs as novel biomarkers for bone diseases—Complex signatures for multifactorial diseases? Mol. Cell. Endocrinol. 2016, 432, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Seeliger, C.; Karpinski, K.; Haug, A.T.; Vester, H.; Schmitt, A.; Bauer, J.S.; van Griensven, M. Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J. Bone Miner. Res. 2014, 29, 1718–1728. [Google Scholar] [CrossRef] [PubMed]
- De-Ugarte, L.; Caro-Molina, E.; Rodriguez-Sanz, M.; Garcia-Perez, M.A.; Olmos, J.M.; Sosa-Henriquez, M.; Perez-Cano, R.; Gomez-Alonso, C.; Del Rio, L.; Mateo-Agudo, J.; et al. SNPs in bone-related miRNAs are associated with the osteoporotic phenotype. Sci. Rep. 2017, 7, 516. [Google Scholar] [CrossRef] [PubMed]
- Kalogeropoulos, M.; Varanasi, S.S.; Olstad, O.K.; Sanderson, P.; Gautvik, V.T.; Reppe, S.; Francis, R.M.; Gautvik, K.M.; Birch, M.A.; Datta, H.K. Zic1 transcription factor in bone: Neural developmental protein regulates mechanotransduction in osteocytes. FASEB J. 2010, 24, 2893–2903. [Google Scholar] [CrossRef] [PubMed]
- Varanasi, S.S.; Olstad, O.K.; Swan, D.C.; Sanderson, P.; Gautvik, V.T.; Reppe, S.; Francis, R.M.; Gautvik, K.M.; Datta, H.K. Skeletal site-related variation in human trabecular bone transcriptome and signaling. PLoS ONE 2010, 5, e10692. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, A.E.; Zheng, T.; Yi, C.; Leaderer, D.; Weidhaas, J.; Slack, F.; Zhang, Y.; Paranjape, T.; Zhu, Y. microRNA miR-196a-2 and breast cancer: A genetic and epigenetic association study and functional analysis. Cancer Res. 2009, 69, 5970–5977. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.S.; Wu, Y.; Zhao, H.G.; Liu, C.X.; Cai, H.Y.; Guo, B.Z.; Xie, Y.A.; Shi, H.R. Association between the rs11614913 variant of miRNA-196a-2 and the risk of epithelial ovarian cancer. Oncol. Lett. 2016, 11, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Haas, U.; Sczakiel, G.; Laufer, S.D. microRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3′-UTR via altered RNA structure. RNA Biol. 2012, 9, 924–937. [Google Scholar] [CrossRef] [PubMed]
- Mahen, E.M.; Watson, P.Y.; Cottrell, J.W.; Fedor, M.J. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010, 8, e1000307. [Google Scholar] [CrossRef] [PubMed]
- Vinci, S.; Gelmini, S.; Pratesi, N.; Conti, S.; Malentacchi, F.; Simi, L.; Pazzagli, M.; Orlando, C. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin. Chem. Lab. Med. 2011, 49, 2073–2080. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Hu, Z.; Xu, Z.; Gu, H.; Yi, L.; Cao, H.; Chen, J.; Tian, T.; Liang, J.; Lin, Y.; et al. Functional variant in microRNA-196a2 contributes to the susceptibility of congenital heart disease in a Chinese population. Hum. Mutat. 2009, 30, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Liu, X.H.; Li, J.H.; Yang, J.S.; Zhang, E.B.; Yin, D.D.; Liu, Z.L.; Zhou, J.; Ding, Y.; Li, S.Q.; et al. miR-196a is upregulated in gastric cancer and promotes cell proliferation by downregulating p27(kip1). Mol. Cancer Ther. 2012, 11, 842–852. [Google Scholar] [CrossRef] [PubMed]
- Zhi, H.; Wang, L.; Ma, G.; Ye, X.; Yu, X.; Zhu, Y.; Zhang, Y.; Zhang, J.; Wang, B. Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin. Res. Cardiol. 2012, 101, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Rao, L.; Peng, Y.; Wang, Y.; Chen, Y.; Song, Y.; Zhang, L. Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin. Chim. Acta 2010, 411, 1287–1290. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.M.; Li, J.; Guo, Y.F.; Cai, F.; Cai, X.X.; Pan, H.Y.; Deng, X.T.; Pan, M. A Functional single-nucleotide polymorphism in pre-microRNA-196a2 is associated with atrial fibrillation in han chinese. Clin. Lab. 2015, 61, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, M.; Sedaghat, S.; de Looper, H.W.; Hofman, A.; Erkeland, S.J.; Franco, O.H.; Dehghan, A. The association of common polymorphisms in miR-196a2 with waist to hip ratio and miR-1908 with serum lipid and glucose. Obesity (Silver Spring) 2015, 23, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Yekta, S.; Shih, I.H.; Bartel, D.P. microRNA-directed cleavage of HOXB8 mRNA. Science 2004, 304, 594–596. [Google Scholar] [CrossRef] [PubMed]
- Alexander, T.; Nolte, C.; Krumlauf, R. Hox genes and segmentation of the hindbrain and axial skeleton. Annu. Rev. Cell Dev. Biol. 2009, 25, 431–456. [Google Scholar] [CrossRef] [PubMed]
- Zakany, J.; Duboule, D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev. 2007, 17, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.; Nakagami, H.; Rodriguez-Araujo, G.; Nimura, K.; Kaneda, Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biol. 2012, 10, e1001314. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Bae, S.W.; Yu, S.S.; Bae, Y.C.; Jung, J.S. miR-196a regulates proliferation and osteogenic differentiation in mesenchymal stem cells derived from human adipose tissue. J. Bone Miner. Res. 2009, 24, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Lei, S.F.; Papasian, C.J.; Deng, H.W. Polymorphisms in predicted miRNA binding sites and osteoporosis. J. Bone Miner. Res. 2011, 26, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Kung, A.W.; Xiao, S.M.; Cherny, S.; Li, G.H.; Gao, Y.; Tso, G.; Lau, K.S.; Luk, K.D.; Liu, J.M.; Cui, B.; et al. Association of JAG1 with bone mineral density and osteoporotic fractures: A genome-wide association study and follow-up replication studies. Am. J. Hum. Genet. 2010, 86, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, I.; Bashinskaya, V.; Kulakova, O.; Baulina, N.; Popova, E.; Boyko, A.; Favorova, O. Variants of microRNA genes: Gender-specific associations with multiple sclerosis risk and severity. Int. J. Mol. Sci. 2015, 16, 20067–20081. [Google Scholar] [CrossRef] [PubMed]
- Hearn, A.P.; Silber, E. Osteoporosis in multiple sclerosis. Mult. Scler. J. 2010, 16, 1031–1043. [Google Scholar] [CrossRef] [PubMed]
- Sioka, C.; Kyritsis, A.P.; Fotopoulos, A. Multiple sclerosis, osteoporosis, and vitamin D. J. Neurol. Sci. 2009, 287, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Gibson, J.C.; Summers, G.D. Bone health in multiple sclerosis. Osteoporos Int. 2011, 22, 2935–2949. [Google Scholar] [CrossRef] [PubMed]
- Estrada, K.; Styrkarsdottir, U.; Evangelou, E.; Hsu, Y.H.; Duncan, E.L.; Ntzani, E.E.; Oei, L.; Albagha, O.M.; Amin, N.; Kemp, J.P.; et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012, 44, 491–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, M.; Ellwanger, D.C.; Hartsperger, M.L.; Pfeufer, A.; Stumpflen, V. Cis-acting polymorphisms affect complex traits through modifications of microRNA regulation pathways. PLoS ONE 2012, 7, e36694. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, M.; Ikram, M.A.; de Looper, H.W.; Hofman, A.; Erkeland, S.J.; Franco, O.H.; Dehghan, A. Genome-wide identification of microRNA-related variants associated with risk of Alzheimer’s disease. Sci. Rep. 2016, 6, 28387. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003, 31, 3429–3431. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Jabbari, H. Sparse RNA folding revisited: Space-efficient minimum free energy structure prediction. Algorithms Mol. Biol. 2016, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Ikram, M.A.; Brusselle, G.G.O.; Murad, S.D.; van Duijn, C.M.; Franco, O.H.; Goedegebure, A.; Klaver, C.C.W.; Nijsten, T.E.C.; Peeters, R.P.; Stricker, B.H.; et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur. J. Epidemiol. 2017, 32, 807–850. [Google Scholar] [CrossRef] [PubMed]
- Watts, N.B.; Adler, R.A.; Bilezikian, J.P.; Drake, M.T.; Eastell, R.; Orwoll, E.S.; Finkelstein, J.S.; Endocrine, S. Osteoporosis in men: An Endocrine Society clinical practice guideline. J. Clin. Endocrinol. Metab. 2012, 97, 1802–1822. [Google Scholar] [CrossRef] [PubMed]
| miRNA ID | Associated Phenotype | Associated Target Genes | p-Value in GWAS Data | Top SNP |
|---|---|---|---|---|
| miR-196a-3p | 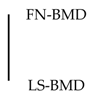 | JAG1 | 1.8 × 10−5 | rs2235811 |
| MACROD2 | 2.0 × 10−6 | rs365824 | ||
| SP1 | 4.2 × 10−5 | rs4759334 | ||
| JAG1 | 4.7 × 10−9 | rs2235811 | ||
| ATF7 | 6.3 × 10−5 | rs1078358 | ||
| MACROD2 | 8.1 × 10−5 | rs6110288 | ||
| miR-196a-5p |  | FRMD4B | 5.6 × 10−4 | rs1564757 |
| NEDD4L | 9.6 × 10−4 | rs533502 | ||
| BIRC6 | 1.2 × 10−3 | rs6737916 | ||
| COL24A1 | 2.6 × 10−3 | rs1359419 | ||
| RSPO2 | 3.1 × 10−3 | rs446454 | ||
| DIP2A | 3.3 × 10−3 | rs2330593 |
| Variables | Men | Women | |
|---|---|---|---|
| FN-BMD (g/cm2) | 0.95 (0.14) | 0.87 (0.14) | |
| LS-BMD (g/cm2) | 1.20 (0.19) | 1.08 (0.19) | |
| Age (years) | 65.71 (10.45) | 66.29 (10.61) | |
| Weight (kg) | 85.55 (12.85) | 73.11 (13.09) | |
| WHR | 0.95 (0.07) | 0.84 (0.07) | |
| Height (cm) | 176.41 (7.01) | 162.73 (6.50) | |
| Alcohol (g/day) | 9.29 (3.57–20.00) | 4.29 (0.54–10.00) | |
| DHEA (nmol/L) | 11.82 (7.32) | 12.31 (7.65) | |
| DHEAS (nmol/L) | 3200.18 (1757.16) | 2099.17 (1337.77) | |
| Androstenedione (nmol/L) | 3.24 (1.27) | 2.70 (1.29) | |
| Testosterone (nmol/L) | 17.53 (5.78) | 0.90 (0.45) | |
| Estradiol (pmol/L) | 96.93 (33.82) | 38.86 (33.18) | |
| Smoking | never smoker | 1125 (42.9%) | 2071 (58.8%) |
| former smoker | 1039 (39.7%) | 841 (23.9%) | |
| current smoker | 456 (17.4%) | 612 (17.4%) | |
| Bone drugs | no | 2607 (99.5%) | 3400 (96.5%) |
| yes | 13 (0.5%) | 124 (3.5%) | |
| Phenotype | Model | Men | Women | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | 95%CI | p-Value | β | 95%CI | p-v | β | 95%CI | p-Value | ||
| FN-BMD | M1 | 0.004 | −0.003, 0.011 | 0.257 | 0.009 | 0.003, 0.014 | 0.003 | 0.007 | 0.003, 0.012 | 0.002 |
| M2 | 0.004 | −0.003, 0.011 | 0.267 | 0.008 | 0.003, 0.014 | 0.003 | 0.007 | 0.003, 0.012 | 0.002 | |
| M3 | 0.004 | −0.004, 0.011 | 0.319 | 0.008 | 0.003, 0.014 | 0.003 | 0.007 | 0.002, 0.011 | 0.003 | |
| LS-BMD | M1 | 0.005 | −0.006, 0.016 | 0.380 | 0.010 | 0.001, 0.019 | 0.023 | 0.009 | 0.002, 0.016 | 0.011 |
| M2 | 0.004 | −0.007, 0.015 | 0.423 | 0.010 | 0.001, 0.018 | 0.026 | 0.009 | 0.002, 0.016 | 0.012 | |
| M3 | 0.003 | −0.008, 0.014 | 0.573 | 0.009 | 0.001, 0.018 | 0.038 | 0.008 | 0.001, 0.015 | 0.020 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karabegović, I.; Maas, S.; Medina-Gomez, C.; Zrimšek, M.; Reppe, S.; Gautvik, K.M.; Uitterlinden, A.G.; Rivadeneira, F.; Ghanbari, M. Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD). Int. J. Mol. Sci. 2017, 18, 2529. https://doi.org/10.3390/ijms18122529
Karabegović I, Maas S, Medina-Gomez C, Zrimšek M, Reppe S, Gautvik KM, Uitterlinden AG, Rivadeneira F, Ghanbari M. Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD). International Journal of Molecular Sciences. 2017; 18(12):2529. https://doi.org/10.3390/ijms18122529
Chicago/Turabian StyleKarabegović, Irma, Silvana Maas, Carolina Medina-Gomez, Maša Zrimšek, Sjur Reppe, Kaare M. Gautvik, André G. Uitterlinden, Fernando Rivadeneira, and Mohsen Ghanbari. 2017. "Genetic Polymorphism of miR-196a-2 is Associated with Bone Mineral Density (BMD)" International Journal of Molecular Sciences 18, no. 12: 2529. https://doi.org/10.3390/ijms18122529





