The POZ/BTB and AT-Hook Containing Zinc Finger 1 (PATZ1) Transcription Regulator: Physiological Functions and Disease Involvement
Abstract
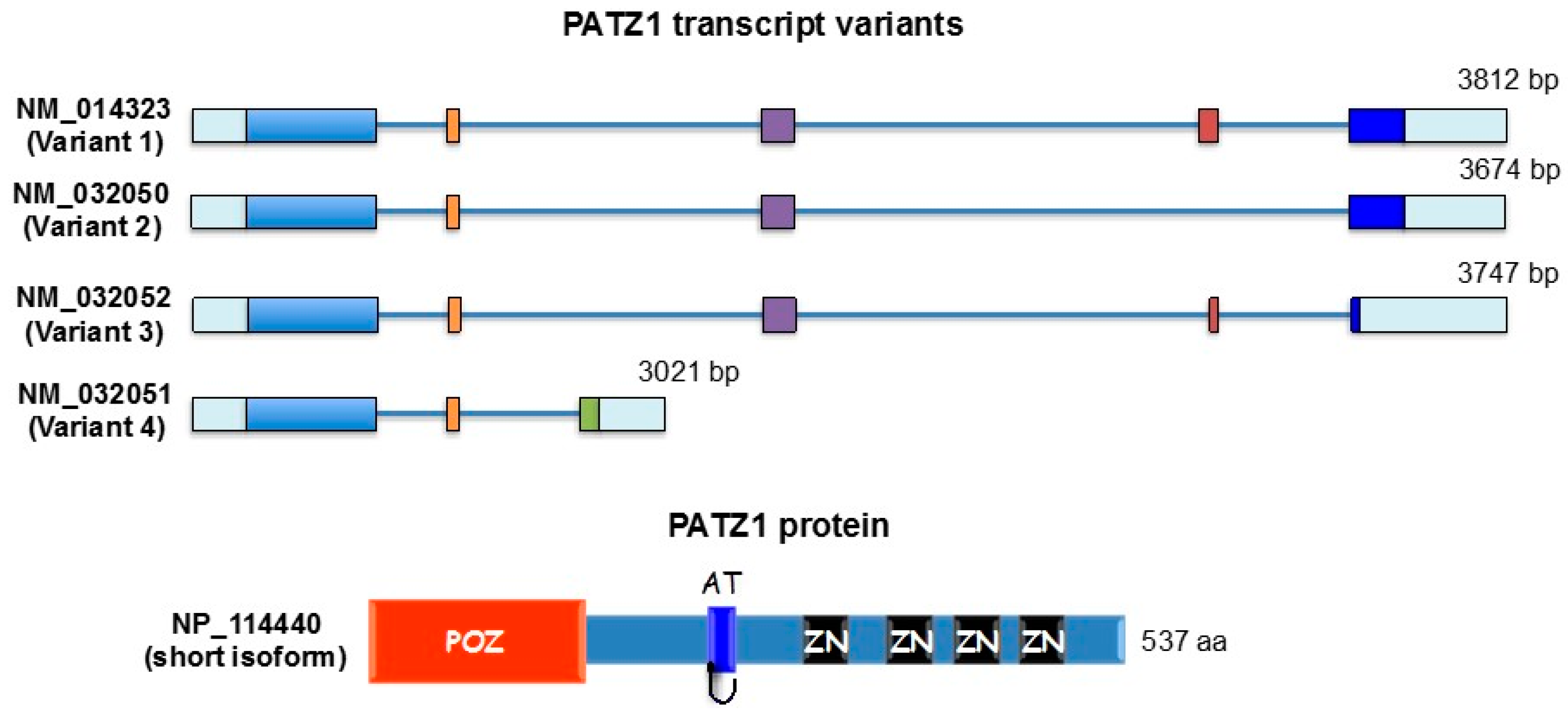
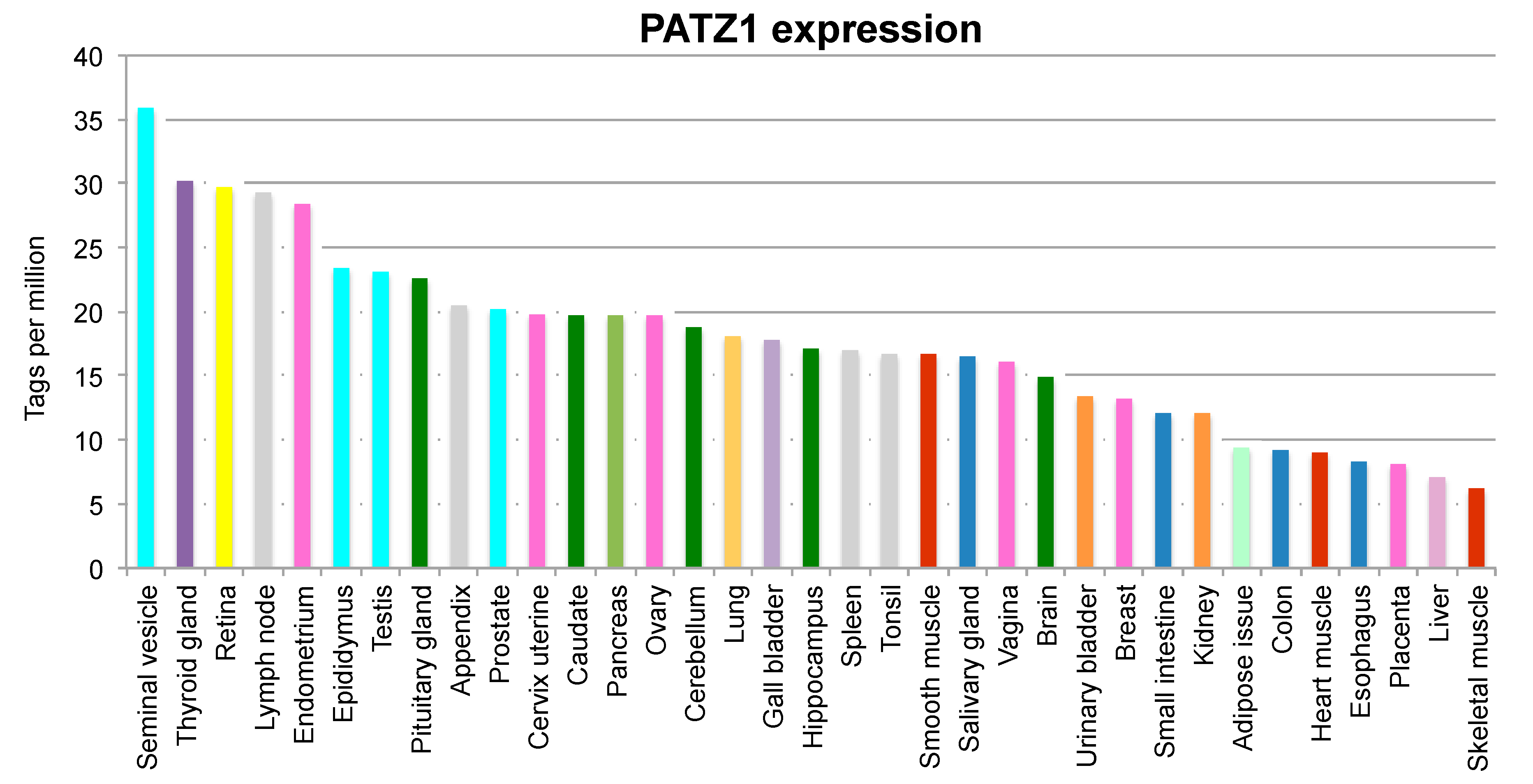
:1. Introduction
2. Physiological Functions of PATZ1 and Pathological Implications
2.1. Cell Proliferation, Senescence and Apoptosis
2.2. Cell Pluripotency and Reprogramming
2.3. Spermatogenesis and Sexual Development
2.4. T Cell Development
3. PATZ1 in Cancer
3.1. Tumor Suppressor Function
3.2. Oncogenic Function
3.3. Double Oncogenic/Tumor Suppressor Function
4. PATZ1 in Other Human Pathologies
5. Final Remarks and Clinical Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| POK | POZ domain Krüppel-like zinc finger |
| CNC | Cap’n’collar |
| bb | Base pairs |
| aa | amino acids |
| IPI | International Proliferation Index |
References
- Valentino, T.; Palmieri, D.; Vitiello, M.; Pierantoni, G.M.; Fusco, A.; Fedele, M. PATZ1 interacts with p53 and regulates expression of p53-target genes enhancing apoptosis or cell survival based on the cellular context. Cell Death Dis. 2013, 4, e963. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Benvenuto, G.; Pero, R.; Majello, B.; Battista, S.; Lembo, F.; Vollono, E.; Day, P.M.; Santoro, M.; Lania, L.; et al. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J. Biol. Chem. 2000, 275, 7894–7901. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Yamagiwa, H.; Hoshino, H.; Muto, A.; Sato, K.; Morita, M.; Hayashi, N.; Yamamoto, M.; Igarashi, K. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol. Cell. Biol. 2000, 20, 1733–1746. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, T.; Modena, P.; Tornielli, S.; Bullrich, F.; Testi, M.A.; Mezzelani, A.; Radice, P.; Azzarelli, A.; Pilotti, S.; Croce, C.M.; et al. A novel zinc finger gene is fused to EWS in small round cell tumor. Oncogene 2000, 19, 3799–3804. [Google Scholar] [CrossRef] [PubMed]
- Klug, A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu. Rev. Biochem. 2010, 79, 213–231. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.F.; Daniel, J.M. POZ for effect—POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006, 16, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Costoya, J.A. Functional analysis of the role of POK transcriptional repressors. Brief. Funct. Genom. Proteom. 2007, 6, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Landsman, D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998, 26, 4413–4421. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Hombauer, M.; Bilic, I.; Naoe, Y.; Schebesta, A.; Taniuchi, I.; Ellmeier, W. The zinc-finger protein MAZR is part of the transcription factor network that controls the CD4 versus CD8 lineage fate of double-positive thymocytes. Nat. Immunol. 2010, 11, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Kim, M.J.; Kim, K.J.; Kim, J.R. POZ/BTB and AT-hook-containing zinc finger protein 1 (PATZ1) inhibits endothelial cell senescence through a p53 dependent pathway. Cell Death Differ. 2012, 19, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, N.; Singh, I.; Mehta, A.; Braun, T.; Barreto, G. HMGA proteins as modulators of chromatin structure during transcriptional activation. Front. Cell Dev. Biol. 2014, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Pierantoni, G.M.; Pallante, P.; Fusco, A. High mobility group A-interacting proteins in cancer: Focus on chromobox protein homolog 7, homeodomain interacting protein kinase 2 and PATZ. J. Nucleic Acids Investig. 2012, 3, 1. [Google Scholar] [CrossRef]
- Pero, R.; Lembo, F.; Palmieri, E.A.; Vitiello, C.; Fedele, M.; Fusco, A.; Bruni, C.B.; Chiariotti, L. PATZ attenuates the RNF4-mediated enhancement of androgen receptor-dependent transcription. J. Biol. Chem. 2002, 277, 3280–3285. [Google Scholar] [CrossRef] [PubMed]
- Keskin, N.; Deniz, E.; Eryilmaz, J.; Un, M.; Batur, T.; Ersahin, T.; Cetin Atalay, R.; Sakaguchi, S.; Ellmeier, W.; Erman, B. PATZ1 Is a DNA Damage-Responsive Transcription Factor That Inhibits p53 Function. Mol. Cell. Biol. 2015, 35, 1741–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, R.; Masuya, M.; Ishii, S.; Katayama, N.; Nosaka, T. Eya2, a Target Activated by Plzf, Is Critical for PLZF-RARA-Induced Leukemogenesis. Mol. Cell. Biol. 2017, 37, e00585-16. [Google Scholar] [CrossRef] [PubMed]
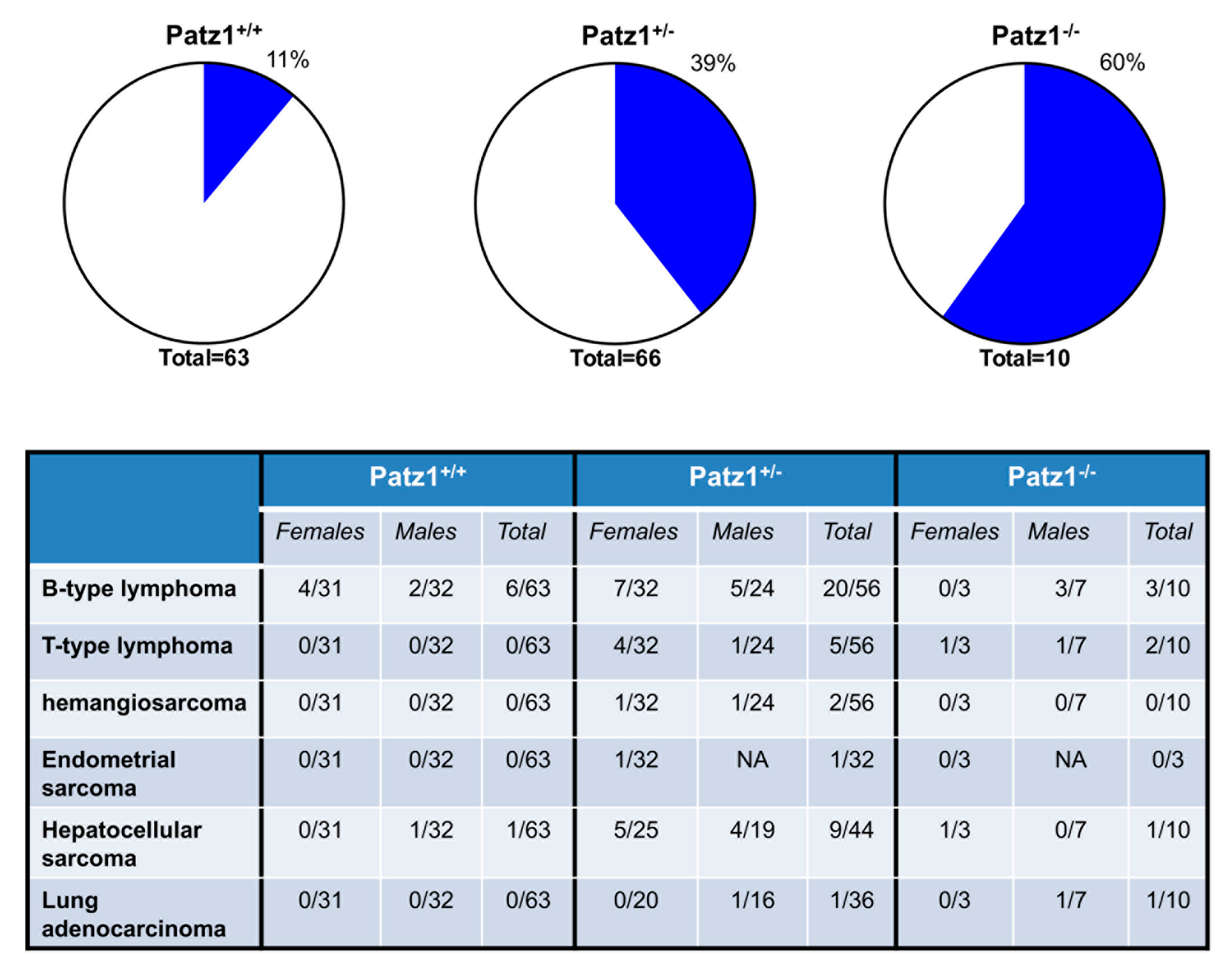
- Valentino, T.; Palmieri, D.; Vitiello, M.; Simeone, A.; Palma, G.; Arra, C.; Chieffi, P.; Chiariotti, L.; Fusco, A.; Fedele, M. Embryonic defects and growth alteration in mice with homozygous disruption of the Patz1 gene. J. Cell. Physiol. 2013, 228, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Franco, R.; Salvatore, G.; Paronetto, M.P.; Barbagallo, F.; Pero, R.; Chiariotti, L.; Sette, C.; Tramontano, D.; Chieffi, G.; et al. PATZ1 gene has a critical role in the spermatogenesis and testicular tumours. J. Pathol. 2008, 215, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.L.; Ravatn, R.; Kudoh, K.; Alabanza, L.; Chin, K.V. Interaction of the regulatory subunit of the cAMP-dependent protein kinase with PATZ1 (ZNF278). Biochem. Biophys. Res. Commun. 2010, 391, 1318–1323. [Google Scholar] [CrossRef] [PubMed]
- Vitiello, M.; Valentino, T.; De Menna, M.; Crescenzi, E.; Francesca, P.; Rea, D.; Arra, C.; Fusco, A.; De Vita, G.; Cerchia, L.; et al. PATZ1 is a target of miR-29b that is induced by Ha-Ras oncogene in rat thyroid cells. Sci. Rep. 2016, 6, 25268. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.X.; Hua, Y.Z.; Wang, N.; Chen, X.; Wang, F.; Liu, J.Y.; Wang, L.L.; Yan, C.Y.; Ma, Y.G.; Cao, Y.H.; et al. miR-24 suppression of POZ/BTB and AT-hook-containing zinc finger protein 1 (PATZ1) protects endothelial cell from diabetic damage. Biochem. Biophys. Res. Commun. 2016, 480, 682–689. [Google Scholar] [CrossRef] [PubMed]
- Ow, J.R.; Ma, H.; Jean, A.; Goh, Z.; Lee, Y.H.; Chong, Y.M.; Soong, R.; Fu, X.Y.; Yang, H.; Wu, Q. Patz1 regulates embryonic stem cell identity. Stem Cells Dev. 2014, 23, 1062–1073. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Lassmann, T.; Murata, M.; Carninci, P. 5′ end-centered expression profiling using cap-analysis gene expression and next-generation sequencing. Nat. Protoc. 2012, 7, 542–561. [Google Scholar] [CrossRef] [PubMed]
- The Human Protein Atlas. Available online: www.proteinatlas.org (accessed on 27 October 2017).
- Schinke, M.; Izumo, S. Deconstructing DiGeorge syndrome. Nat. Genet. 2001, 27, 238–240. [Google Scholar] [CrossRef] [PubMed]
- Liguori, G.; Mancinelli, S.; CNR-IGB, Naples, Italy; Vitiello, M.; Fedele, M.; CNR-IEOS, Naples, Italy. Personal Communication, 2017.
- Ma, H.; Ow, J.R.; Tan, B.C.; Goh, Z.; Feng, B.; Loh, Y.H.; Fedele, M.; Li, H.; Wu, Q. The dosage of Patz1 modulates reprogramming process. Sci. Rep. 2014, 4, 7519. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Segura, A.; de Jong, T.V.; Melov, S.; Guryev, V.; Campisi, J.; Demaria, M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr. Biol. 2017, 27, 2652–2660. [Google Scholar] [CrossRef] [PubMed]
- Tritz, R.; Mueller, B.M.; Hickey, M.J.; Lin, A.H.; Gomez, G.G.; Hadwiger, P.; Sah, D.W.; Muldoon, L.; Neuwelt, E.A.; Kruse, C.A. siRNA Down-regulation of the PATZ1 Gene in Human Glioma Cells Increases Their Sensitivity to Apoptotic Stimuli. Cancer Ther. 2008, 6, 865–876. [Google Scholar] [PubMed]
- Oliviero, G.; Munawar, N.; Watson, A.; Streubel, G.; Manning, G.; Bardwell, V.; Bracken, A.P.; Cagney, G. The variant Polycomb Repressor Complex 1 component PCGF1 interacts with a pluripotency sub-network that includes DPPA4, a regulator of embryogenesis. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, A.; Xin, L.; Sharov, A.A.; Thomas, M.; Mowrer, G.; Meyers, E.; Piao, Y.; Mehta, S.; Yee, S.; Nakatake, Y.; et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell 2009, 5, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Guadagno, E.; Vitiello, M.; Francesca, P.; Calì, G.; Caponnetto, F.; Cesselli, D.; Camorani, S.; Borrelli, G.; Califano, M.; Cappabianca, P.; et al. PATZ1 is a new prognostic marker of glioblastoma associated with the stem-like phenotype and enriched in the proneural subtype. Oncotarget 2017, 8, 59282–59300. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Hara, L.; Smith, L.B. Androgen receptor roles in spermatogenesis and infertility. Best Pract. Res. Clin. Endocrinol. Metab. 2015, 29, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Cousminer, D.L.; Stergiakouli, E.; Berry, D.J.; Ang, W.; Groen-Blokhuis, M.M.; Körner, A.; Siitonen, N.; Ntalla, I.; Marinelli, M.; Perry, J.R.; et al. Genome-wide association study of sexual maturation in males and females highlights a role for body mass and menarche loci in male puberty. Hum. Mol. Genet. 2014, 23, 4452–4464. [Google Scholar] [CrossRef] [PubMed]
- Bilic, I.; Koesters, C.; Unger, B.; Sekimata, M.; Hertweck, A.; Maschek, R.; Wilson, C.B.; Ellmeier, W. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat. Immunol. 2006, 7, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, S.; Hainberger, D.; Tizian, C.; Tanaka, H.; Okuda, T.; Taniuchi, I.; Ellmeier, W. MAZR and Runx Factors Synergistically Repress ThPOK during CD8+ T Cell Lineage Development. J. Immunol. 2015, 195, 2879–2887. [Google Scholar] [CrossRef] [PubMed]
- Ellmeier, W. Molecular control of CD4+ T cell lineage plasticity and integrity. Int. Immunopharmacol. 2015, 28, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Burrow, A.A.; Williams, L.E.; Pierce, L.C.; Wang, Y.H. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genom. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Klein-Szanto, A.J.; Fox Chase Cancer Center, Philadelphia, PA, USA; Franco, R.; University of Naples L. Vanvitelli, Naples, Italy; Fedele, M.; CNR-IEOS, Naples, Italy. Personal Communication, 2010.
- Chiappetta, G.; Valentino, T.; Vitiello, M.; Pasquinelli, R.; Monaco, M.; Palma, G.; Sepe, R.; Luciano, A.; Pallante, P.; Palmieri, D.; et al. PATZ1 acts as a tumor suppressor in thyroid cancer via targeting p53-dependent genes involved in EMT and cell migration. Oncotarget 2015, 6, 5310–5323. [Google Scholar] [CrossRef] [PubMed]
- Franco, R.; Scognamiglio, G.; Valentino, E.; Vitiello, M.; Luciano, A.; Palma, G.; Arra, C.; La Mantia, E.; Panico, L.; Tenneriello, V.; et al. PATZ1 expression correlates positively with BAX and negatively with BCL6 and survival in human diffuse large B cell lymphomas. Oncotarget 2016, 7, 59158–59172. [Google Scholar] [CrossRef] [PubMed]
- Francesca, P. Oncogenic Ras Downregulates PATZ1 during Transformation of Rat Thyroid Cells to Enhance Their Proliferation, Migration and Growth Capacity of Stem-Like Thyrospheres. Magistral Thesis, Medical Biotechnologies, University of Naples Federico II, Naples, Italy, 2015. [Google Scholar]
- Ho, M.Y.; Liang, C.M.; Liang, S.M. PATZ1 induces PP4R2 to form a negative feedback loop on IKK/NF-κB signaling in lung cancer. Oncotarget 2016, 7, 52255–52269. [Google Scholar] [CrossRef] [PubMed]
- Fedele, M.; Cerchia, L.; Chiappetta, G. The Tumor Suppressive role of PATZ1 in Thyroid Cancer: A matter of Epithelial-Mesenchymal Transition. Chemother. Open Access 2016, 5. [Google Scholar] [CrossRef]
- Esposito, F.; Boscia, F.; Franco, R.; Tornincasa, M.; Fusco, A.; Kitazawa, S.; Looijenga, L.H.; Chieffi, P. Down-regulation of oestrogen receptor-β associates with transcriptional co-regulator PATZ1 delocalization in human testicular seminomas. J. Pathol. 2011, 224, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Görnemann, J.; Hofmann, T.G.; Will, H.; Müller, M. Interaction of human papillomavirus type 16 L2 with cellular proteins: Identification of novel nuclear body-associated proteins. Virology 2002, 303, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Dobbelstein, M.; Strano, S.; Roth, J.; Blandino, G. p73-induced apoptosis: A question of compartments and cooperation. Biochem. Biophys. Res. Commun. 2005, 331, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Sun, D.; Zhang, Y.; Zhao, S.; Xiong, H.; Fang, J. Zinc finger protein 278, a potential oncogene in human colorectal cancer. Acta Biochim. Biophys. Sin. 2008, 40, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.J.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 2683–2710. [Google Scholar] [CrossRef] [PubMed]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Severson, E.; Gay, L.; Vergilio, J.A.; Elvin, J.; Suh, J.; Daniel, S.; Covert, M.; Frampton, G.M.; Hsu, S.; et al. Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 2017, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.A.; Sun, X.M.; Yan, C.Y.; Liu, L.; Hao, M.W.; Liu, Q.; Jiao, X.Y.; Liang, Y.M. Hyperglycemia-induced PATZ1 negatively modulates endothelial vasculogenesis via repression of FABP4 signaling. Biochem. Biophys. Res. Commun. 2016, 477, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Dhaouadi, N.; Li, J.Y.; Feugier, P.; Gustin, M.P.; Dab, H.; Kacem, K.; Bricca, G.; Cerutti, C. Computational identification of potential transcriptional regulators of TGF-β1 in human atherosclerotic arteries. Genomics 2014, 103, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yang, J.; Chen, J.; Wu, Q.; Gong, W.; Zhang, J.; Shao, W.; Mu, J.; Yang, D.; Yang, Y.; et al. Differential co-expression and regulation analyses reveal different mechanisms underlying major depressive disorder and subsyndromal symptomatic depression. BMC Bioinform. 2015, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Han, N.; Li, Z.; Lu, Q. Identification of transcription regulatory relationships in rheumatoid arthritis and osteoarthritis. Clin. Rheumatol. 2013, 32, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Eliwa, H.; Belzung, C.; Surget, A. Adult hippocampal neurogenesis: Is it the alpha and omega of antidepressant action? Biochem. Pharmacol. 2017, 141, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Ameur, A.; Enroth, S.; Bysani, M.; Nord, H.; Cavalli, M.; Essand, M.; Gyllensten, U.; Wadelius, C. PATZ1 down-regulates FADS1 by binding to rs174557 and is opposed by SP1/SREBP1c. Nucleic Acids Res. 2017, 45, 2408–2422. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Wang, Q.; Zhang, W.; Bian, A.; Zhang, J. Identification of genes associated with renal cell carcinoma using gene expression profiling analysis. Oncol. Lett. 2016, 12, 73–78. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fedele, M.; Crescenzi, E.; Cerchia, L. The POZ/BTB and AT-Hook Containing Zinc Finger 1 (PATZ1) Transcription Regulator: Physiological Functions and Disease Involvement. Int. J. Mol. Sci. 2017, 18, 2524. https://doi.org/10.3390/ijms18122524
Fedele M, Crescenzi E, Cerchia L. The POZ/BTB and AT-Hook Containing Zinc Finger 1 (PATZ1) Transcription Regulator: Physiological Functions and Disease Involvement. International Journal of Molecular Sciences. 2017; 18(12):2524. https://doi.org/10.3390/ijms18122524
Chicago/Turabian StyleFedele, Monica, Elvira Crescenzi, and Laura Cerchia. 2017. "The POZ/BTB and AT-Hook Containing Zinc Finger 1 (PATZ1) Transcription Regulator: Physiological Functions and Disease Involvement" International Journal of Molecular Sciences 18, no. 12: 2524. https://doi.org/10.3390/ijms18122524





