Exploration for the Salinity Tolerance-Related Genes from Xero-Halophyte Atriplex canescens Exploiting Yeast Functional Screening System
Abstract
:1. Introduction
2. Results
2.1. Generation of the Yeast Expression cDNA Library
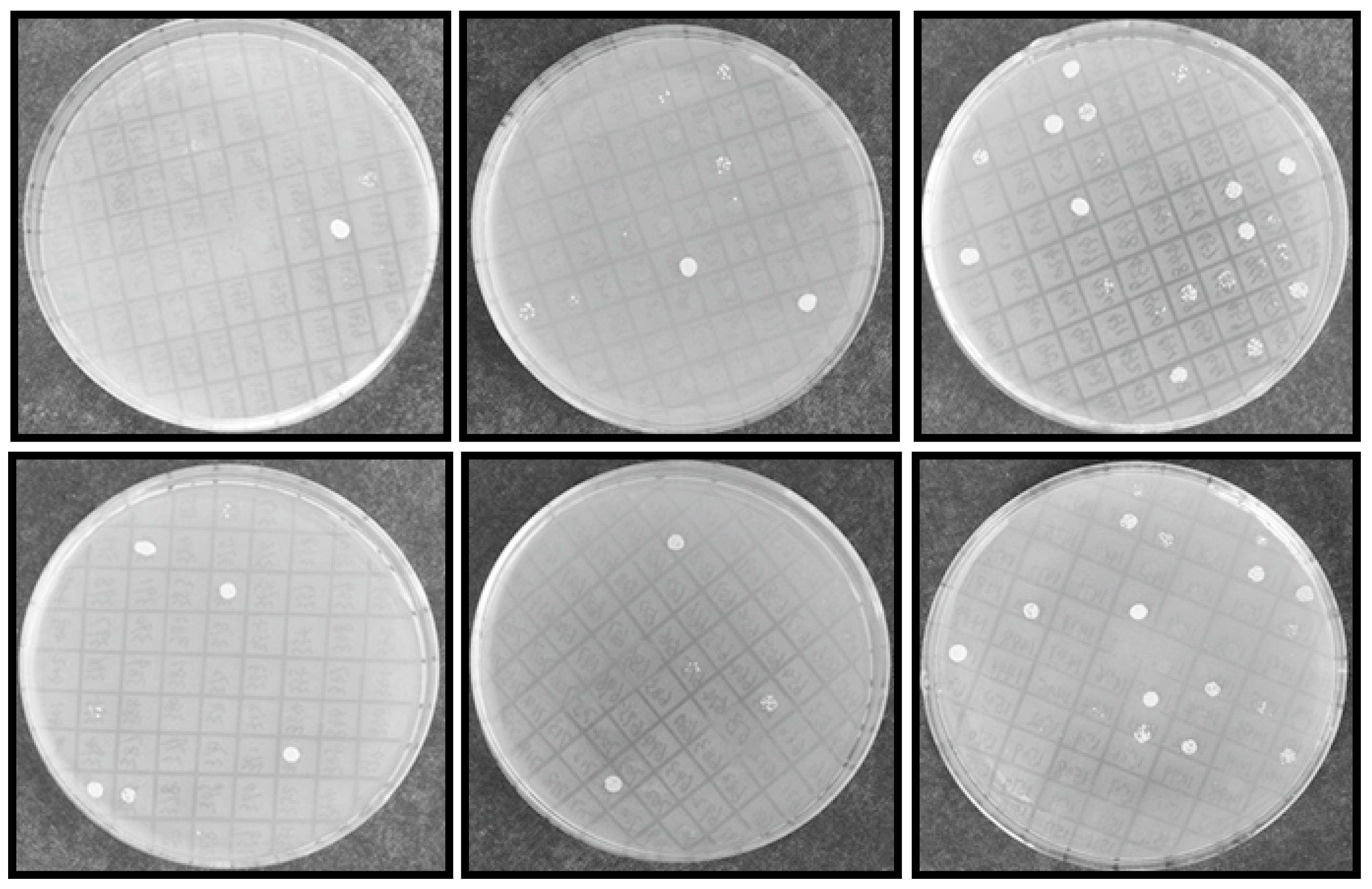
2.2. Identification of Salt Tolerance-Related Genes from A. canescens
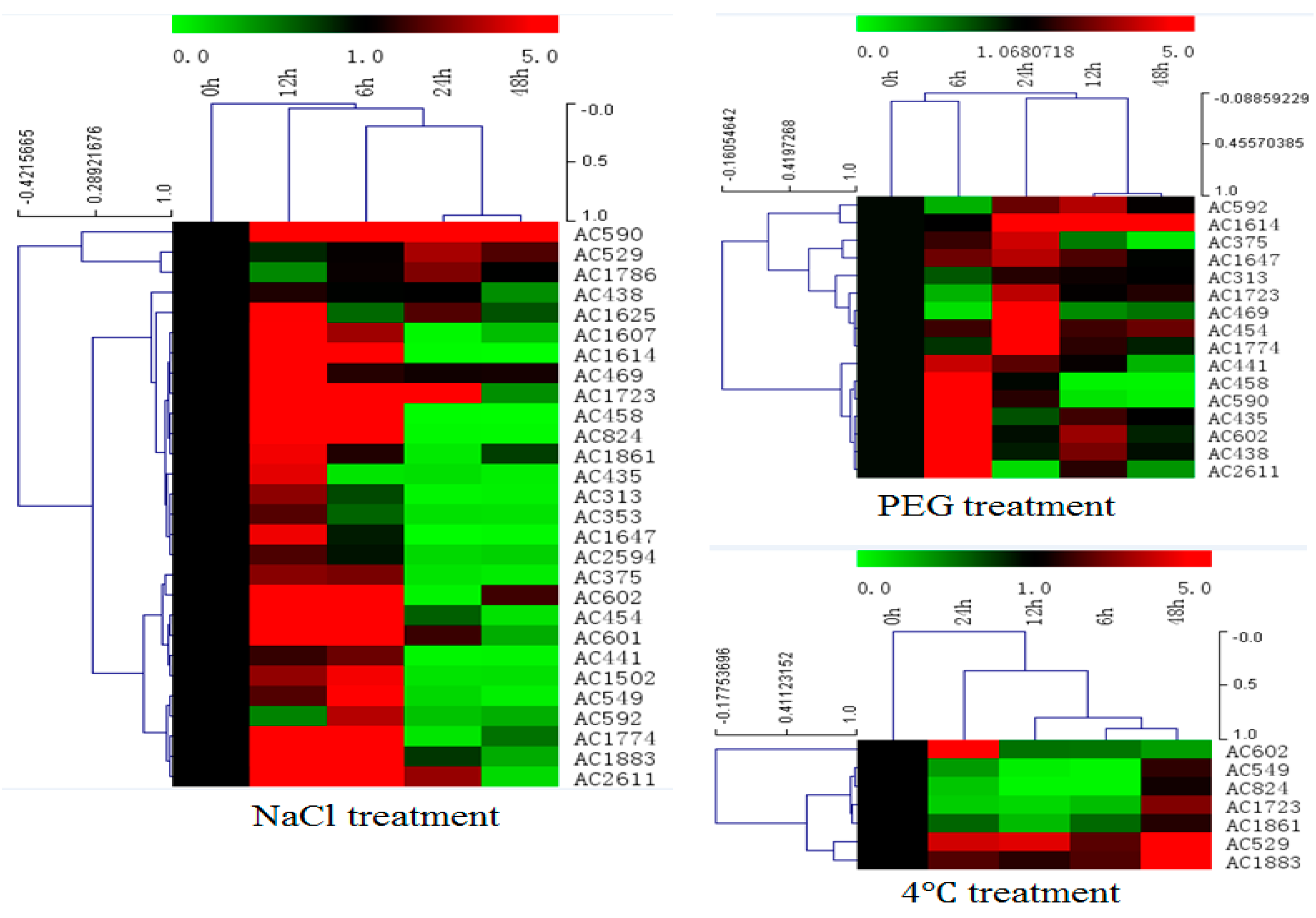
2.3. Expression Profiles of Selected Salt Resistance Genes under Abiotic Stress
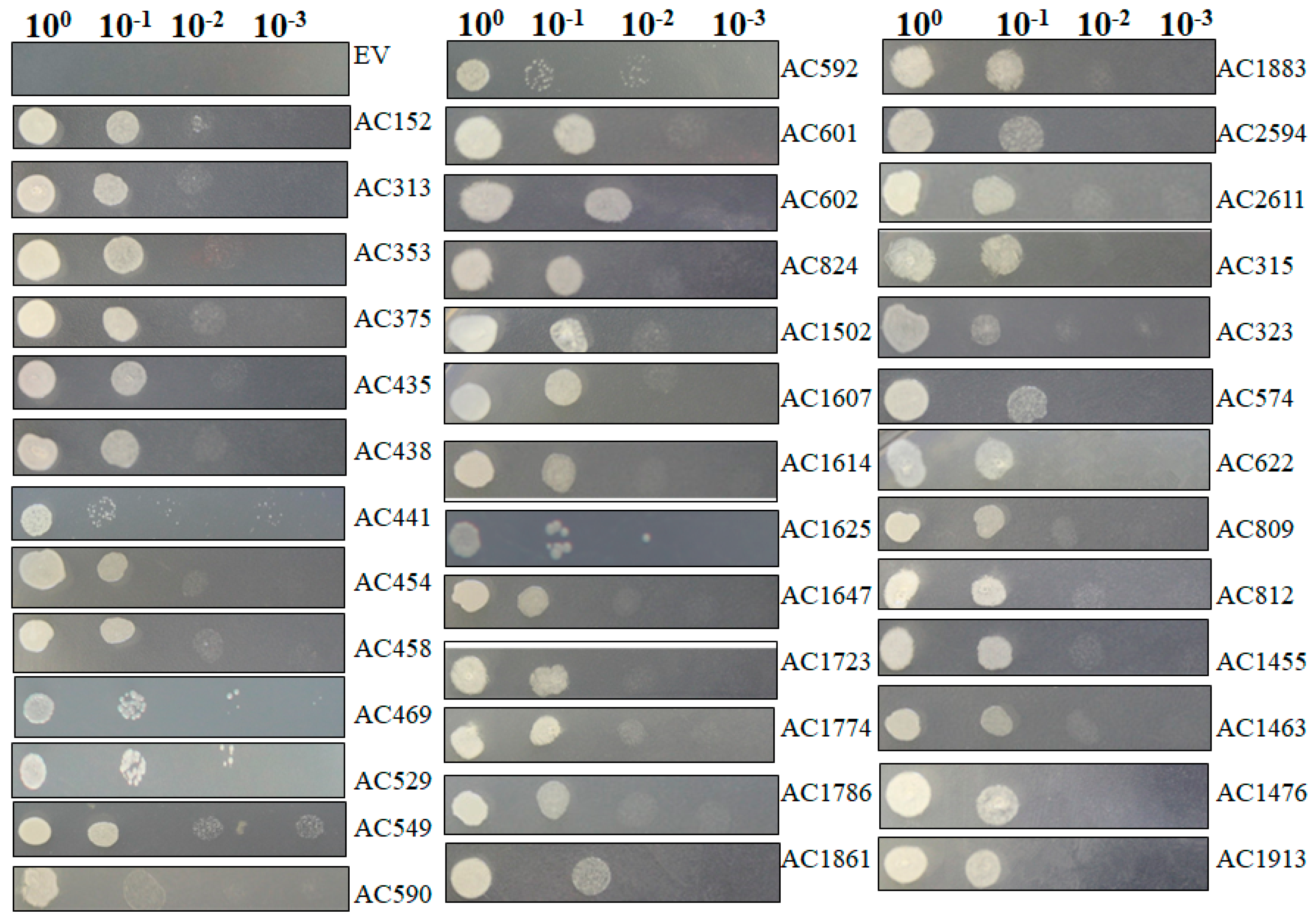
2.4. Recapturing Salt Resistance Assay of Isolated Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Plasmid, Yeast and Escherichia coli Strains, Medium
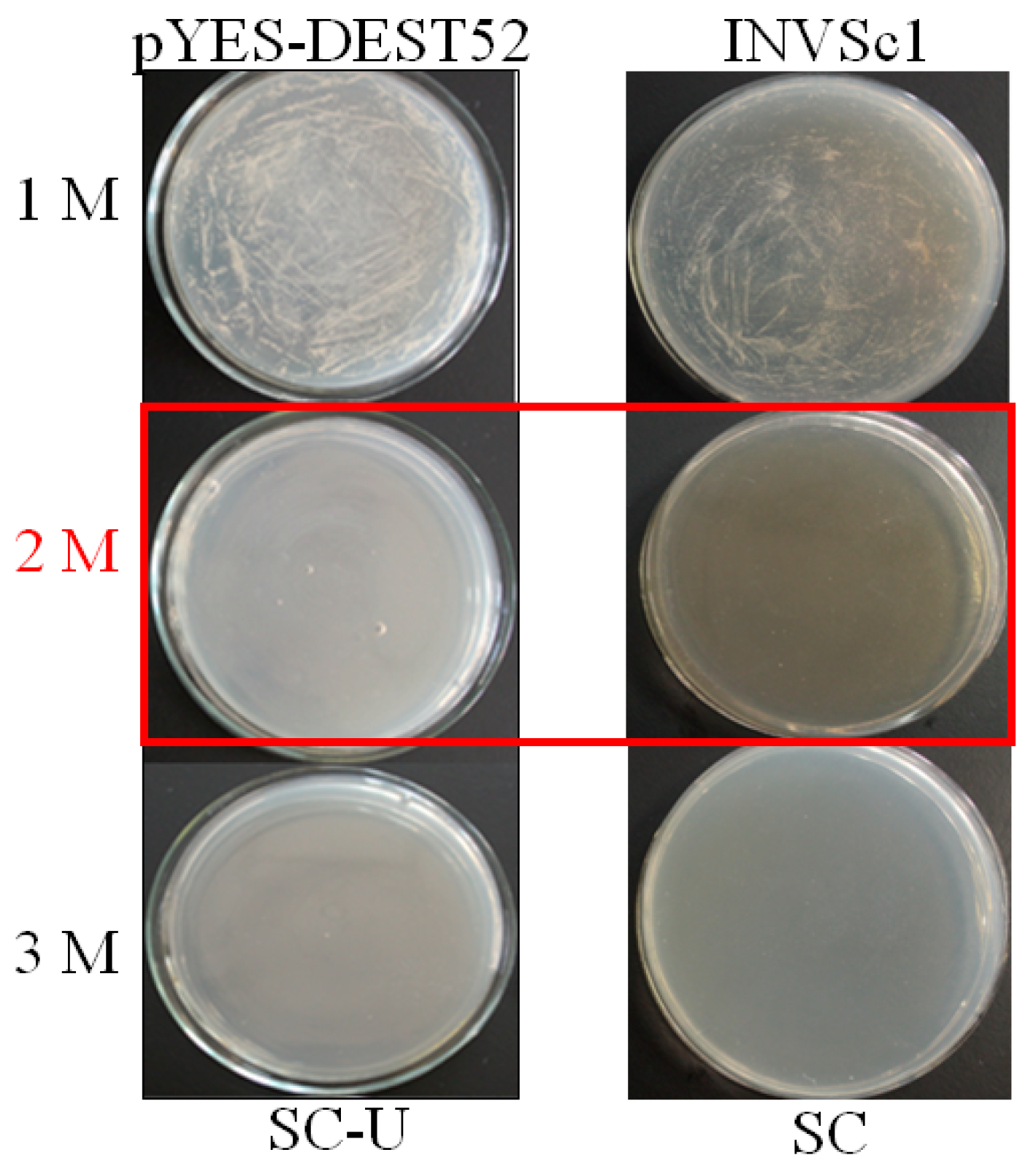
4.2. Yeast Functional Screening to Select the Salinity Tolerant Transformants
4.3. Plasmid Rescue, Sequencing and Data Analysis
4.4. Functional Annotation and Categorization
4.5. Quantitative RT-PCR Validation of Salt-Related Genes
4.6. Salt-Stress Assay for Isolated Salt Resistance Genes
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant Cellular and Molecular Responses to High Salinity. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Tanna, B. Halophytes: Potential Resources for Salt Stress Tolerance Genes and Promoters. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Jorge, T.F.; Duro, N.; da Costa, M.; Florian, A.; Ramalho, J.C.; Ribeiro-Barros, A.I.; Fernie, A.R.; António, C. GC-TOF-MS analysis reveals salt stress-responsive primary metabolites in Casuarina glauca tissues. Metabolomics 2017, 13, 95. [Google Scholar] [CrossRef]
- Scotti-Campos, P.; Duro, N.; Costa, M.D.; Pais, I.P.; Rodrigues, A.P.; Batista-Santos, P.; Semedo, J.N.; Leitão, A.E.; Lidon, F.C.; Pawlowski, K.; et al. Antioxidative ability and membrane integrity in salt-induced responses of Casuarina glauca Sieber ex Spreng. in symbiosis with N2-fixing Frankia Thr or supplemented with mineral nitrogen. J. Plant Physiol. 2016, 196–197, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Duro, N.; Batista-Santos, P.; da Costa, M.; Maia, R.; Castro, I.V.; Ramos, M.; Ramalho, J.C.; Pawlowski, K.; Máguas, C.; Ribeiro-Barros, A. The impact of salinity on the symbiosis between Casuarina glauca Sieb. ex Spreng. and N2-fixing Frankia bacteria based on the analysis of Nitrogen and Carbon metabolism. Plant Soil 2016, 398, 327–337. [Google Scholar] [CrossRef]
- Batista-Santos, P.; Duro, N.; Rodrigues, A.P.; Semedo, J.N.; Alves, P.; da Costa, M.; Graça, I.; Pais, I.P.; Scotti-Campos, P.; Lidon, F.C.; et al. Is salt stress tolerance in Casuarina glauca Sieb. ex Spreng. associated with its nitrogen-fixing root-nodule symbiosis? An analysis at the photosynthetic level. Plant Physiol. Biochem. 2015, 96, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Meng, Y.; Li, B.; Ma, X.; Lai, Y.; Si, E.; Yang, K.E.; Xu, X.; Shang, X.; Wang, H.; et al. Physiological and proteomic analyses of salt stress response in the halophyte Halogeton glomeratus. Plant Cell Environ. 2015, 38, 655–669. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S. Learning from halophytes: Physiological basis and strategies to improve abiotic stress tolerance in crops. Ann. Bot. 2013, 112, 1209–1221. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-J.; Zhang, Z.; Wang, J.-Y.; Oh, D.-H.; Dassanayake, M.; Liu, B.; Huang, Q.; Sun, H.-X.; Xia, R.; Wu, Y.; et al. Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. USA 2012, 109, 12219–12224. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yin, H.; Yang, L.; Xie, Z.; Liu, X. Differential salt tolerance in seedlings derived from dimorphic seeds of Atriplex centralasiatica: From physiology to molecular analysis. Planta 2011, 233, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, M.-W.; Xie, M.; Liu, X.; Ni, M.; Shao, G.; Song, C.; Kay-Yuen Yim, A.; Tao, Y.; Wong, F.-L.; et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Benzarti, M.; Ben Rejeb, K.; Debez, A.; Abdelly, C. Environmental and economical opportunities for the valorisation of the genus Atriplex: New insights. In Crop Improvement: New Approaches and Modern Techniques; Hakeem, R.K., Ahmad, P., Ozturk, M., Eds.; Springer: Boston, MA, USA, 2013. [Google Scholar]
- Buckholz, R.G.; Gleeson, M.A.G. Yeast systems for the commercial production of heterologous proteins. Nat. Biotechnol. 1991, 9, 1067–1072. [Google Scholar] [CrossRef]
- Nakahara, Y.; Sawabe, S.; Kainuma, K.; Katsuhara, M.; Shibasaka, M.; Suzuki, M.; Yamamoto, K.; Oguri, S.; Sakamoto, H. Yeast functional screen to identify genes conferring salt stress tolerance in Salicornia europaea. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Kappachery, S.; Yu, J.W.; Baniekal-Hiremath, G.; Park, S.W. Rapid identification of potential drought tolerance genes from Solanum tuberosum by using a yeast functional screening method. Comptes Rendus Biol. 2013, 336, 530–545. [Google Scholar] [CrossRef] [PubMed]
- Gangadhar, B.; Yu, J.; Sajeesh, K.; Park, S. A systematic exploration of high-temperature stress-responsive genes in potato using large-scale yeast functional screening. Mol. Genet. Genom. 2013, 289, 185–201. [Google Scholar] [CrossRef] [PubMed]
- Eswaran, N.; Parameswaran, S.; Sathram, B.; Anantharaman, B.; Kumar, G.R.; Tangirala, S. Yeast functional screen to identify genetic determinants capable of conferring abiotic stress tolerance in Jatropha curcas. BMC Biotechnol. 2010, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, X.; Yu, G.; Jia, C.; Liu, J.; Pan, H. Generation and analysis of expressed sequence tags (ESTs) from halophyte Atriplex canescens to explore salt-responsive related genes. Int. J. Mol. Sci. 2014, 15, 11172–11189. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Plant salt tolerance. Trends Plant Sci. 2001, 6, 66–71. [Google Scholar] [CrossRef]
- Sun, X.-H.; Yu, G.; Li, J.-T.; Jia, P.; Zhang, J.-C.; Jia, C.-G.; Zhang, Y.-H.; Pan, H.-Y. A heavy metal-associated protein (AcHMA1) from the halophyte, Atriplex canescens (Pursh) Nutt., confers tolerance to iron and other abiotic stresses when expressed in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2014, 15, 14891–14906. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, J.; Sun, X.; Zhang, X.; Liu, J.; Pan, H. Overexpression of AcNIP5;1, a novel nodulin-like intrinsic protein from halophyte Atriplex canescens, enhances sensitivity to salinity and improves drought tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2015, 33, 1864–1875. [Google Scholar] [CrossRef]
- Li, J.; Yu, G.; Sun, X.; Liu, Y.; Liu, J.; Zhang, X.; Jia, C.; Pan, H. AcPIP2, a plasma membrane intrinsic protein from halophyte Atriplex canescens, enhances plant growth rate and abiotic stress tolerance when overexpressed in Arabidopsis thaliana. Plant Cell Rep. 2015, 34, 1401–1415. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, G.; Sun, X.; Zhang, X.; Liu, J.; Pan, H. AcEBP1, an ErbB3-Binding Protein (EBP1) from halophyte Atriplex canescens, negatively regulates cell growth and stress responses in Arabidopsis. Plant Sci. 2016, 248, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.-Q.; Guo, H.; Wang, S.-M.; Zhao, B.; Zhang, J.-L.; Ma, Q.; Yin, H.-J.; Bao, A.-K. The photosynthesis, Na+/K+ homeostasis and osmotic adjustment of Atriplex canescens in response to salinity. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-D.; Xie, L.; Wei, Y.; Zhou, X.; Jia, B.; Liu, J.; Zhang, S. Abiotic Stress Resistance, a Novel Moonlighting Function of Ribosomal Protein RPL44 in the Halophilic Fungus Aspergillus glaucus. Appl. Environ. Microbiol. 2014, 80, 4294–4300. [Google Scholar] [CrossRef] [PubMed]
- Agarie, S.; Shimoda, T.; Shimizu, Y.; Baumann, K.; Sunagawa, H.; Kondo, A.; Ueno, O.; Nakahara, T.; Nose, A.; Cushman, J.C. Salt tolerance, salt accumulation, and ionic homeostasis in an epidermal bladder-cell-less mutant of the common ice plant Mesembryanthemum crystallinum. J. Exp. Bot. 2007, 58, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Casas, A.M.; Nelson, D.E.; Raghothama, K.G.; D’Urzo, M.P.; Singh, N.K.; Bressan, R.A.; Hasegawa, P.M. Expression of Osmotin-Like Genes in the Halophyte Atriplex nummularia L. Plant Physiol. 1992, 99, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Yamaji, N. A cooperative system of silicon transport in plants. Trends Plant Sci. 2015, 20, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Ballesta, M.D.C.; Carvajal, M. New challenges in plant aquaporin biotechnology. Plant Sci. 2014, 217–218, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Lecourieux, F.; Kappel, C.; Lecourieux, D.; Serrano, A.; Torres, E.; Arce-Johnson, P.; Delrot, S. An update on sugar transport and signalling in grapevine. J. Exp. Bot. 2014, 65, 821–832. [Google Scholar] [CrossRef] [PubMed]
- Sugihara, K.; Hanagata, N.; Dubinsky, Z.; Baba, S.; Karube, I. Molecular Characterization of cDNA Encoding Oxygen Evolving Enhancer Protein 1 Increased by Salt Treatment in the Mangrove Bruguiera gymnorrhiza. Plant Cell Physiol. 2000, 41, 1279–1285. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jia, P.; Zhang, J.; Pan, H. Cloning and characterization of a salt responsive gene AcPsbQ1 from Atriplex canescens. Hereditas (Beijing) 2015, 37, 84–90. [Google Scholar]
- Zhou, W.; Jia, C.-G.; Wu, X.; Hu, R.-X.; Yu, G.; Zhang, X.-H.; Liu, J.-L.; Pan, H.-Y. ZmDBF3, a Novel Transcription Factor from Maize (Zea mays L.), Is Involved in Multiple Abiotic Stress Tolerance. Plant Mol. Biol. Rep. 2015, 34, 353–364. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.; Li, H.; Wang, Y.; Zhang, Y.; Wood, A.J. EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 2014, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, D.; Li, X.; Guan, K.; Yang, H. Novel DREB A-5 subgroup transcription factors from desert moss (Syntrichia caninervis) confers multiple abiotic stress tolerance to yeast. J. Plant Physiol. 2016, 194, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Romanos, M.A.; Scorer, C.A.; Clare, J.J. Foreign gene expression in yeast: A review. Yeast 1992, 8, 423–488. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
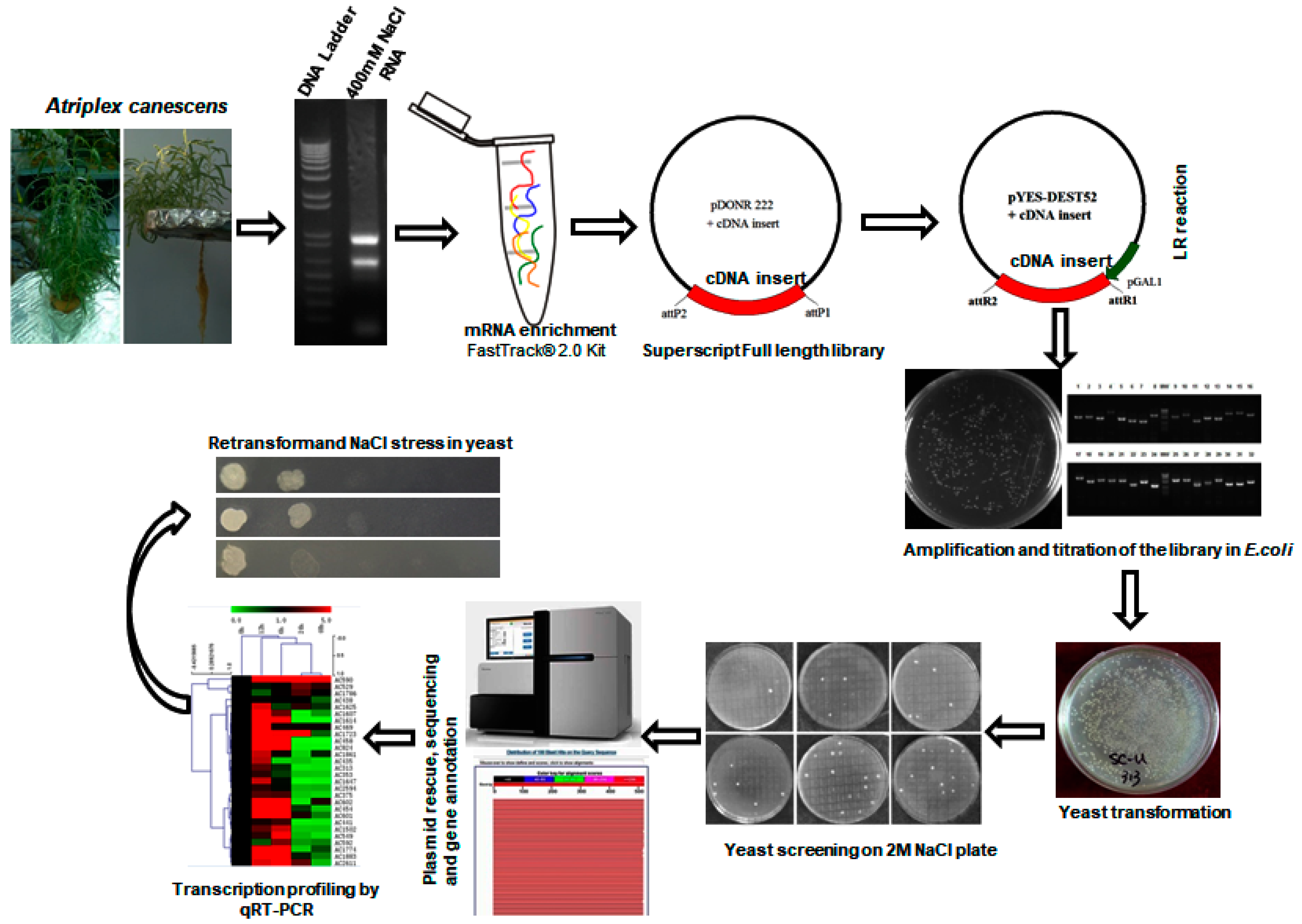

| cDNA library | Vector | CFU | Average Inserted Fragment | Recombinant Rate |
|---|---|---|---|---|
| Primary cDNA library | pDONR222 | 1.76 × 106 | >1 kb | 91% |
| Destination cDNA library | pYES-DEST52 | 1.624 × 107 | >1 kb | 95% |
| Clone No. | GenBank Accession No. | Putative Gene Function | Organism Matched | Full-Length ORF | E-Value | Target GenBank Accession No. | Recapturing Salt Resistance |
|---|---|---|---|---|---|---|---|
| Ac113 | KJ026988 | Cysteine desulfurase | Arabidopsis thaliana | Yes | 2 × 10−70 | NP_001078802.1 | No |
| Ac151 | KJ026991 | Phosphoglyceride transfer family protein | Theobroma cacao | Yes | 5 × 10−160 | EOY17339.1 | No |
| Ac152 | KJ026992 | Cyclophilin | Suaeda salsa | Yes | 5 × 10−92 | AGI78541.1 | Yes |
| Ac264 | KJ026993 | Phosphorylase superfamily protein | Theobroma cacao | Yes | 5 × 10−40 | EOY10097.1 | No |
| Ac273 | KJ026995 | Beta-amylase | Ricinus communis | Yes | 3 × 10−21 | XP_002519919.1 | No |
| Ac313 | KJ026998 | Membrane-associated progesterone binding protein | Theobroma cacao | Yes | 4 × 10−46 | EOY29287.1 | Yes |
| Ac315 | KJ027000 | Signal recognition particle receptor | Theobroma cacao | Yes | 7 × 10−122 | EOY25233.1 | Yes |
| Ac323 | KJ027001 | Pre-mRNA cleavage complex II protein Clp1 | Zea mays | No | 9 × 10−113 | AFW66051.1 | Yes |
| Ac353 | KJ027002 | K+ uptake permease 7 | Theobroma cacao | No | 2 × 10−119 | EOX98796.1 | Yes |
| Ac354 | KJ027003 | Chlorophyll a/b-binding protein | Solanum lycopersicum | Yes | 8 × 10−144 | AAA34140.1 | No |
| Ac359 | KJ027004 | Cytochrome c1-1 | Solanum lycopersicum | Yes | 2 × 10−174 | XP_004231797.1 | No |
| Ac375 | KJ027006 | Glycogen synthase kinase-3 | Ricinus communis | No | 2 × 10−149 | XP_002522231.1 | Yes |
| Ac392 | KJ027007 | Major chlorophyll a/b binding protein LHCb1.2 | Spinacia oleracea | Yes | 0 | CAJ77390.1 | No |
| Ac435 | KJ027013 | 3-ketoacyl-CoA thiolase 2, peroxisomal-like | Solanum lycopersicum | Yes | 0 | XP_004247828.1 | Yes |
| Ac438 | KJ027014 | Glycine-rich protein | Pisum sativum | Yes | 6 × 10−3 | CAH40798.1 | Yes |
| Ac441 | KJ027018 | Zinc metalloprotease | Ricinus communis | No | 4 × 10−63 | XP_002518787.1 | Yes |
| Ac454 | KJ027022 | RNA-binding family protein with retrovirus zinc finger-like domain | Theobroma cacao | Yes | 2 × 10−66 | EOY18432.1 | Yes |
| Ac458 | KJ027023 | Cytochrome P450 | Populus trichocarpa | Yes | 1 × 10−104 | XP_002334834.1 | Yes |
| Ac469 | KJ027025 | Oxygen-evolving enhancer protein 3 | Spinacia oleracea | Yes | 8 × 10−130 | P12301.1 | Yes |
| Ac529 | KJ027035 | Temperature-induced lipocalin | Populus balsamifera | Yes | 2 × 10−80 | ABB02389.1 | Yes |
| Ac549 | KJ027036 | Late embryogenesis abundant protein | Ricinus communis | Yes | 5 × 10−07 | XP_002509455.1 | Yes |
| Ac567 | KJ027038 | Polyubiquitin family protein | Populus trichocarpa | No | 0 | XP_006372519.1 | No |
| Ac574 | KJ027042 | Tetratricopeptide repeat (TPR)-like superfamily protein | Theobroma cacao | Yes | 2 × 10−104 | EOY31556.1 | Yes |
| Ac590 | KJ027045 | Ubiquitin | Arabidopsis thaliana | No | 4 × 10−55 | ABH08753.1 | Yes |
| Ac592 | KJ027046 | Zinc finger (C3HC4-type RING finger) family protein | Arabidopsis thaliana | Yes | 1 × 10−94 | NP_564060.1 | Yes |
| Ac601 | KJ027047 | ARF domain class transcription factor | Malus domestica | No | 4 × 10−88 | ADL36578.1 | Yes |
| Ac603 | KJ027048 | Dehydration-responsive element binding protein | Atriplex canescens | Yes | 0 | AEW68339.1 | No |
| Ac602 | KJ027049 | Cysteine proteinase A494 | Populus trichocarpa | Yes | 0 | XP_002305451.2 | Yes |
| Ac622 | KJ027051 | AP2-like ethylene-responsive transcription factor | Medicago truncatula | No | 1 × 10−63 | XP_003625138.1 | Yes |
| Ac799 | KJ027055 | S-adenosyl-L-homocysteine hydrolase | Beta vulgaris | Yes | 0 | BAE07182.1 | No |
| Ac809 | KJ027057 | Alanine aminotransferase 2 | Theobroma cacao | Yes | 3 × 10−113 | EOY27390.1 | Yes |
| Ac812 | KJ027058 | Armadillo/beta-catenin repeat family protein | Theobroma cacao | Yes | 3 × 10−115 | EOY18778.1 | Yes |
| Ac824 | KJ027061 | Hexose transporter | Elaeis guineensis | Yes | 8 × 10−123 | AEQ94177.1 | Yes |
| Ac1455 | KJ027067 | Zinc finger and BTB domain-containing protein | Theobroma cacao | No | 2 × 10−61 | EOY17025.1 | Yes |
| Ac1458 | KJ027069 | Thiamin biosynthetic enzyme | Glycine max | Yes | 0 | BAA88226.1 | No |
| Ac1463 | KJ027070 | GTP-binding protein sar1 | Ricinus communis | No | 1 × 10−121 | XP_002515297.1 | Yes |
| Ac1476 | KJ027073 | Nonspecific lipid-transfer protein-like protein At2g13820-like | Vitis vinifera | No | 9 × 10−35 | XP_003632312.1 | Yes |
| Ac1502 | KJ027076 | NAD(P)H dehydrogenase C1 isoform 2 | Theobroma cacao | No | 7 × 10−153 | EOX92707.1 | Yes |
| Ac1607 | KJ027083 | Profilin | Chenopodium album | No | 6 × 10−83 | Q84V37.1 | Yes |
| Ac1614 | KJ027084 | Leucine-rich repeat protein kinase family protein | Arabidopsis thaliana | No | 3 × 10−27 | NP_190219.1 | Yes |
| Ac1625 | KJ027085 | eukaryotic elongation factor 1A | Salsola komarovii | No | 2 × 10−160 | BAC22127.1 | Yes |
| Ac1637 | KJ027087 | Ferredoxin-1 | Mesembryanthemum crystallinum | No | 4 × 10−66 | O04683.1 | No |
| Ac1647 | KJ027088 | RNA-binding family protein | Theobroma cacao | Yes | 4 × 10−71 | EOY04889.1 | Yes |
| Ac1723 | KJ027090 | Late embryogenesis abundant protein D-113 | Gossypium hirsutum | No | 1 × 10−6 | P09441.2 | Yes |
| Ac1737 | KJ027095 | Glyceraldehyde-3-phosphate dehydrogenase | Atriplex nummularia | No | 0 | P34783.1 | No |
| Ac1752 | KJ027097 | Putative ankyrin-repeat protein | Vitis aestivalis | No | 4 × 10−104 | AAQ96339.1 | No |
| Ac1774 | KJ027102 | Cysteine proteinases | Theobroma cacao | Yes | 4 × 10−115 | EOX95504.1 | Yes |
| Ac1786 | KJ027105 | Calcium-dependent lipid-binding (CaLB domain) plant phosphoribosyltransferase | Theobroma cacao | Yes | 0 | EOY09444.1 | Yes |
| Ac1861 | KJ027110 | calmodulin1 | Zea mays | Yes | 5 × 10−98 | AFW78488.1 | Yes |
| Ac1883 | KJ027112 | Abscisic acid stress ripening protein | Mesembryanthemum crystallinum | Yes | 9 × 10−5 | AAC14177.1 | Yes |
| Ac1913 | KJ027114 | Non-specific lipid-transfer protein | Beta vulgaris | Yes | 2 × 10−24 | Q43748.1 | Yes |
| Ac2594 | KJ027115 | Aquaporin NIP6.1 family protein | Populus trichocarpa | Yes | 6 × 10−135 | XP_002304723.1 | Yes |
| Ac2611 | KJ027117 | 23 kDa precursor protein of the oxygen-evolving complex | Salicornia europaea | Yes | 4 × 10−155 | BAG70022.1 | Yes |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, G.; Li, J.; Sun, X.; Liu, Y.; Wang, X.; Zhang, H.; Pan, H. Exploration for the Salinity Tolerance-Related Genes from Xero-Halophyte Atriplex canescens Exploiting Yeast Functional Screening System. Int. J. Mol. Sci. 2017, 18, 2444. https://doi.org/10.3390/ijms18112444
Yu G, Li J, Sun X, Liu Y, Wang X, Zhang H, Pan H. Exploration for the Salinity Tolerance-Related Genes from Xero-Halophyte Atriplex canescens Exploiting Yeast Functional Screening System. International Journal of Molecular Sciences. 2017; 18(11):2444. https://doi.org/10.3390/ijms18112444
Chicago/Turabian StyleYu, Gang, Jingtao Li, Xinhua Sun, Yanzhi Liu, Xueliang Wang, Hao Zhang, and Hongyu Pan. 2017. "Exploration for the Salinity Tolerance-Related Genes from Xero-Halophyte Atriplex canescens Exploiting Yeast Functional Screening System" International Journal of Molecular Sciences 18, no. 11: 2444. https://doi.org/10.3390/ijms18112444





