The Role of Non-Coding RNAs in Cytoplasmic Male Sterility in Flowering Plants
Abstract
:1. Introduction
2. Mitochondrial CMS Genes and Their Mode of Action
3. Restoration of Fertility by Nuclear Genes
4. Non-Coding RNAs in Pollen Development and CMS
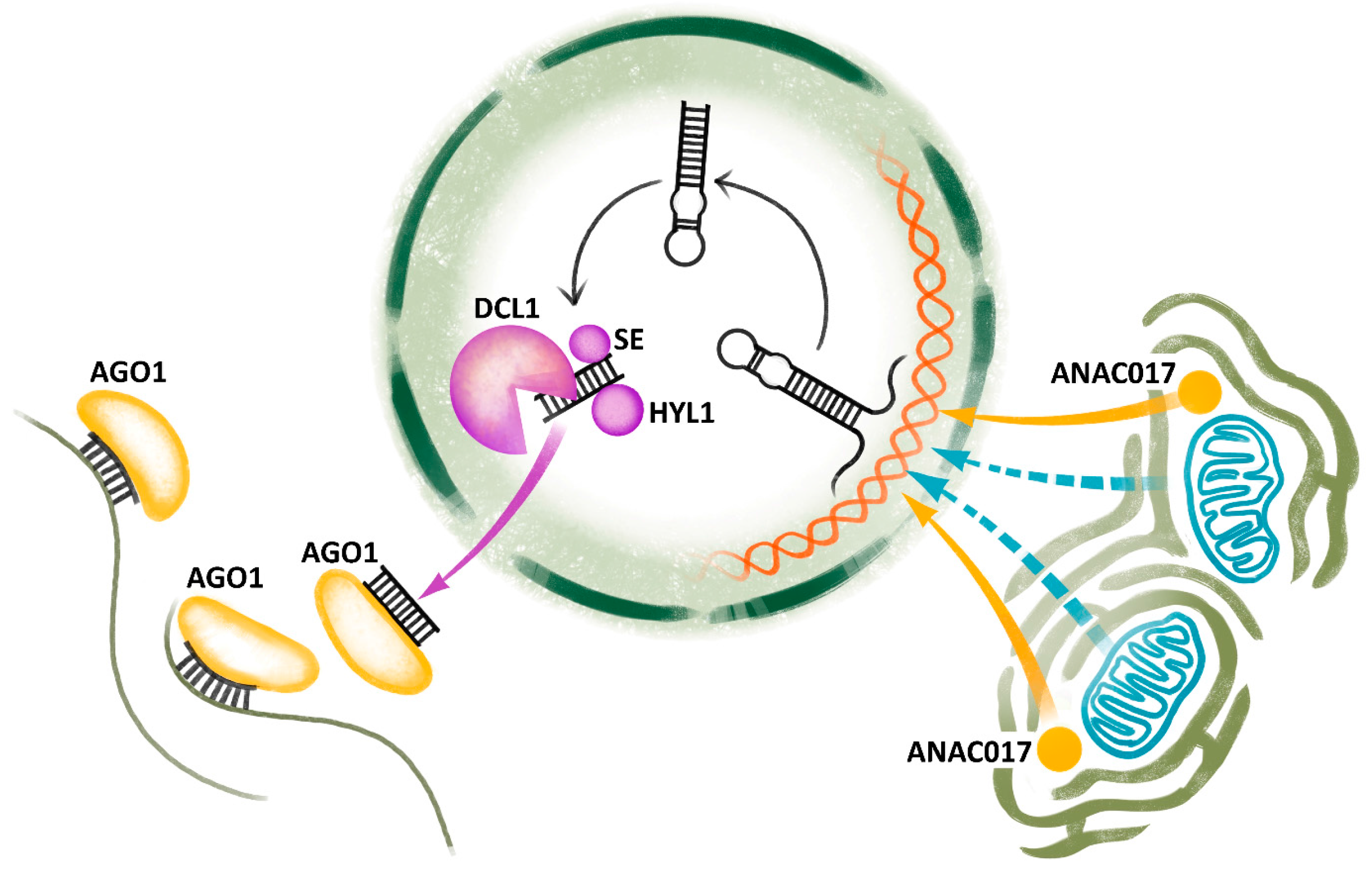
5. miRNAs
6. siRNAs and ta-si RNAs
7. lncRNA
8. Non-Coding RNAs Encoded by the Mitochondrial Genome
9. Future Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, R.; Marine, J.C.; Leucci, E. Non-coding RNAs: The dark side of nuclear-mitochondrial communication. EMBO J. 2017, 36, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Touzet, P.; Meyer, E.H. Cytoplasmic male sterility and mitochondrial metabolism in plants. Mitochondrion 2014, 19, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Hanson, M.R.; Bentolila, S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 2004, 16, S154–S169. [Google Scholar] [CrossRef] [PubMed]
- Gillman, J.D.; Bentolila, S.; Hanson, M.R. The petunia restorer of fertility protein is part of a large mitochondrial complex that interacts with transcripts of the CMS-associated locus. Plant J. 2007, 49, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.H.; Kang, J.G.; Kim, B.D. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol. Biol. 2007, 63, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Duroc, Y.; Hiard, S.; Vrielynck, N.; Ragu, S.; Budar, F. The Ogura sterility-inducing protein forms a large complex without interfering with the oxidative phosphorylation components in rapeseed mitochondria. Plant Mol. Biol. 2009, 70, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Dewey, R.E.; Timothy, D.H.; Levings, C.S. A mitochondrial protein associated with cytoplasmic male-sterility in the T-cytoplasm of maize. Proc. Natl. Acad. Sci. USA 1987, 84, 5374–5378. [Google Scholar] [CrossRef] [PubMed]
- Ducos, E.; Touzet, P.; Boutry, M. The male sterile G cytoplasm of wild beet displays modified mitochondrial respiratory complexes. Plant J. 2001, 26, 171–180. [Google Scholar] [CrossRef]
- Itabashi, E.; Kazama, T.; Toriyama, K. Characterization of cytoplasmic male sterility of rice with Lead Rice cytoplasm in comparison with that with Chinsurah Boro II cytoplasm. Plant Cell Rep. 2009, 28, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gao, F.; Ji, Y.X.; Liu, Y.; Dan, Z.W.; Yang, P.F.; Zhu, Y.G.; Li, S.Q. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol. 2013, 198, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Zabala, G.; Gabay-Laughnan, S.; Laughnan, J.R. The nuclear gene Rf3 affects the expression of the mitochondrial chimeric sequence R implicated in S-type male sterility in maize. Genetics 1997, 147, 847–860. [Google Scholar] [PubMed]
- Yamamoto, M.P.; Kubo, T.; Mikami, T. The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol. Genet. Genom. 2005, 273, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Grelon, M.; Budar, F.; Bonhomme, S.; Pelletier, G. Ogura cytoplasmic male sterility (CMS)-associated ORF138 is translated into a mitochondrial-membrane polypeptide in male-sterile Brassica cybrids. Mol. Gen. Genet. 1994, 243, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Kazama, T.; Itabashi, E.; Fujii, S.; Nakamura, T.; Toriyama, K. Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J. 2016, 85, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Balk, J.; Leaver, C.J. The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 2001, 13, 1803–1818. [Google Scholar] [CrossRef] [PubMed]
- Twell, D. Male gametogenesis and germline specification in flowering plants. Sex. Plant Reprod. 2011, 24, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.P.; Xu, H.; Liu, Z.L.; Guo, J.X.; Li, H.Y.; Chen, L.T.; Fang, C.; Zhang, Q.Y.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Shafiq, S.; Li, J.R.; Sun, Q.W. Functions of plants long non-coding RNAs. Biochim. Biophys. Acta 2016, 1859, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Wise, R.P. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998, 3, 175–180. [Google Scholar] [CrossRef]
- Sabar, M.; Gagliardi, D.; Balk, J.; Leaver, C.J. ORFB is a subunit of F1FO-ATP synthase: Insight into the basis of cytoplasmic male sterility in sunflower. EMBO Rep. 2003, 4, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Kazama, T.; Nakamura, T.; Watanabe, M.; Sugita, M.; Toriyama, K. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008, 55, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Mitra, J.; Bhattacharyya, J.; Pradhan, S.; Sikdar, N.; Das, S.; Chakraborty, S.; Kumar, S.; Lakhanpaul, S.; Sen, S.K. Transgenic expression of an unedited mitochondrial orfB gene product from wild abortive (WA) cytoplasm of rice (Oryza sativa L.) generates male sterility in fertile rice lines. Planta 2015, 241, 1463–1479. [Google Scholar] [CrossRef] [PubMed]
- Uyttewaal, M.; Arnal, N.; Quadrado, M.; Martin-Canadell, A.; Vrielynck, N.; Hiard, S.; Gherbi, H.; Bendahmane, A.; Budar, F.; Mireau, H. Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 2008, 20, 3331–3345. [Google Scholar] [CrossRef] [PubMed]
- Sarria, R.; Lyznik, A.; Vallejos, C.E.; Mackenzie, S.A. A cytoplasmic male sterility-associated mitochondrial peptide in common bean is post-translationally regulated. Plant Cell 1998, 10, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Gaborieau, L.; Brown, G.G.; Mireau, H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci. 2016, 7, 1816. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Haertel, B.; Brennicke, A. RNA Editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Bond, C.S.; Small, I.D. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc. Natl. Acad. Sci. USA 2011, 108, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Melonek, J.; Stone, J.D.; Small, I. Evolutionary plasticity of restorer-of-fertility-like proteins in rice. Sci. Rep. 2016, 6, 35152. [Google Scholar] [CrossRef] [PubMed]
- Gouyon, P.H.; Couvet, D. A conflict between two sexes, females and hermaphrodites. In The Evolution of Sex and Its Consequences; Stearns, S.C., Ed.; Birkhauser Verlag: Basel, Switzerland, 1987; pp. 245–261. ISBN 978-3-0348-6273-8. [Google Scholar]
- Cui, X.; Wise, R.P.; Schnable, P.S. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 1996, 272, 1334–1336. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Cui, X.; Horner, H.T.; Weiner, H.; Schnable, P.S. Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. Plant Cell 2001, 13, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Toriyama, K. Suppressed expression of RETROGRADE-REGULATED MALE STERILITY restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. USA 2009, 106, 9513–9518. [Google Scholar] [CrossRef] [PubMed]
- McCauley, D.E.; Olson, M.S. Do recent findings in plant mitochondrial molecular and population genetics have implications for the study of gynodioecy and cytonuclear conflict? Evolution 2008, 62, 1013–1025. [Google Scholar] [CrossRef] [PubMed]
- Case, A.L.; Willis, J.H. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution 2008, 62, 1026–1039. [Google Scholar] [CrossRef] [PubMed]
- Darracq, A.; Varré, J.S.; Marechal-Drouard, L.; Courseaux, A.; Castric, V.; Saumitou-Laprade, P.; Oztas, S.; Lenoble, P.; Vacherie, B.; Barbe, V.; et al. Structural and content diversity of mitochondrial genome in beet: A comparative genomic analysis. Genome Biol. Evol. 2011, 3, 723–736. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Case, A.L.; Floro, E.R.; Willis, J.H. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 2012, 4, 670–686. [Google Scholar] [CrossRef] [PubMed]
- Štorchová, H.; Müller, K.; Lau, S.; Olson, M.S. Mosaic origin of a complex chimeric mitochondrial gene in Silene vulgaris. PLoS ONE 2012, 7, e30401. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Yang, J.; Pan, F.; Jin, B. Differential expression of microRNAs may regulate pollen development in Brassica oleracea. Genet. Mol. Res. 2015, 14, 15024–15034. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.D.; Štorchová, H. The application of RNA-seq to the comprehensive analysis of plant mitochondrial transcriptomes. Mol. Genet. Genom. 2015, 290, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.J.; Zhang, H.Y.; Zheng, Y.Z.; Ding, Y. Comparative expression profiling of miRNAs between the cytoplasmic male sterile line MeixiangA and its maintainer line MeixiangB during rice anther development. Planta 2015, 241, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xie, Y.; Xu, L.; Wang, Y.; Zhu, X.W.; Wang, R.H.; Zhang, Y.; Muleke, E.M.; Liu, L.W. Identification of microRNAs and Their Target Genes Explores miRNA-Mediated Regulatory Network of Cytoplasmic Male Sterility Occurrence during Anther Development in Radish (Raphanus sativus L.). Front. Plant Sci. 2016, 7, 1054. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.K.; Li, Y.; Zhang, C.W.; Duan, W.K.; Huang, F.Y.; Hou, X.L. Basic helix-loop-helix transcription factor BcbHLHpol functions as a positive regulator of pollen development in non-heading Chinese cabbage. Funct. Integr. Genom. 2014, 14, 731–739. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.W.; Ding, X.L.; Wang, X.; He, T.T.; Zhang, H.; Yang, L.S.; Wang, T.L.; Chen, L.F.; Gai, J.Y.; Yang, S.P. Genome-wide comparative analysis of DNA methylation between soybean cytoplasmic male-sterile line NJCMS5A and its maintainer NJCMS5B. BMC Genom. 2017, 18, 596. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Achkar, N.P.; Cambiagno, D.A.; Manavella, P.A. miRNA biogenesis: A dynamic pathway. Trends Plant Sci. 2016, 21, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Castillo-Gonzalez, C.; Yu, B.; Zhang, X.R. The functions of plant small RNAs in development and in stress responses. Plant J. 2017, 90, 654–670. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.; Voinnet, O.; Chappell, L.; Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 2002, 21, 4671–4679. [Google Scholar] [CrossRef] [PubMed]
- Zilberman, D.; Cao, X.F.; Jacobsen, S.E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 2003, 299, 716–719. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, K.B.; Guang, S.; Buckley, B.A.; Wong, L.; Bochner, A.F.; Kennedy, S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011, 7, e1002249. [Google Scholar] [CrossRef] [PubMed]
- MacLean, D.; Elina, N.; Havecker, E.R.; Heimstaedt, S.B.; Studholme, D.J.; Baulcombe, D.C. Evidence for Large Complex Networks of Plant Short Silencing RNAs. PLoS ONE 2010, 5, e9901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamthan, A.; Chaudhuri, A.; Kamthan, M.; Datta, A. Small RNAs in plants: Recent development and application for crop improvement. Front. Plant Sci. 2015, 6, 208. [Google Scholar] [CrossRef]
- Rogers, K.; Chen, X.M. Biogenesis, Turnover, and Mode of Action of Plant MicroRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; You, C.J.; Chen, X.M. The evolution of microRNAs in plants. Curr. Opin. Plant Biol. 2017, 35, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Y.; Wu, M.; Li, L.H.; Jin, C.; Zhang, Q.L.; Chen, C.B.; Song, W.Q.; Wang, C.G. Small RNA Sequencing Reveals Differential miRNA Expression in the Early Development of Broccoli (Brassica oleracea var. italica) Pollen. Front. Plant Sci. 2017, 8, 404. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.H.; Liu, X.Y.; Xu, B.C.; Zhao, N.; Yang, X.D.; Zhang, M.F. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genom. 2013, 14, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.N.; Zheng, B.B.; Wang, L.; Wu, X.M.; Xu, Q.; Guo, W.W. High-throughput sequencing and degradome analysis reveal altered expression of miRNAs and their targets in a male-sterile cybrid pummelo (Citrus grandis). BMC Genom. 2016, 17, 591. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.C.; Zhang, X.H.; Yao, Q.J.; Yuan, Y.X.; Li, X.X.; We, F.; Zhao, Y.Y.; Zhang, Q.; Wang, Z.Y.; Jiang, W.S.; et al. The miRNAs and their regulatory networks responsible for pollen abortion in Ogura-CMS Chinese cabbage revealed by high-throughput sequencing of miRNAs, degradomes, and transcriptomes. Front. Plant Sci. 2015, 6, 894. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.L.; Li, J.J.; Zhang, H.; He, T.T.; Han, S.H.; Li, Y.W.; Yang, S.P.; Gai, J.Y. Identification of miRNAs and their targets by high-throughput sequencing and degradome analysis in cytoplasmic male-sterile line NJCMS1A and its maintainer NJCMS1B of soybean. BMC Genom. 2016, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, Z.M.; Lin, H.J.; Liu, H.J.; Chen, J.; Peng, H.; Cao, M.J.; Rong, T.Z.; Pan, G.T. Cytoplasmic male sterility-regulated novel microRNAs from maize. Funct. Integr. Genom. 2011, 11, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Han, S.H.; Ding, X.L.; He, T.T.; Dai, J.Y.; Yang, S.P.; Gai, J.Y. Comparative Transcriptome Analysis between the Cytoplasmic Male Sterile Line NJCMS1A and Its Maintainer NJCMS1B in Soybean (Glycine max (L.) Merr.). PLoS ONE 2015, 10, e0126771. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Yang, S.P.; Gai, J.Y. Transcriptome comparative analysis between the cytoplasmic male sterile line and fertile line in soybean (Glycine max (L.) Merr.). Genes Genom. 2017, 39, 1117–1127. [Google Scholar] [CrossRef]
- Ng, S.; Ivanova, A.; Duncan, O.; Law, S.R.; van Aken, O.; de Clercq, I.; Wang, Y.; Carrie, C.; Xu, L.; Kmiec, B.; et al. A membrane-Bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 2013, 25, 3450–3471. [Google Scholar] [CrossRef] [PubMed]
- Van Aken, O.; Ford, E.; Lister, R.; Huang, S.B.; Millar, A.H. Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 2016, 88, 542–558. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.X.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Peragine, A.; Park, M.Y.; Poethig, R.S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005, 19, 2164–2175. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.D.; Fahlgren, N.; Chapman, E.J.; Cumbie, J.S.; Sullivan, C.M.; Givan, S.A.; Kasschau, K.D.; Carrington, J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell 2007, 19, 926–942. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Meyers, B.C.; Liu, Z.C.; Beers, E.P.; Ye, S.Q.; Liu, Z.R. MicroRNA Superfamilies Descended from miR390 and Their Roles in Secondary Small Interfering RNA Biogenesis in Eudicots. Plant Cell 2013, 25, 1555–1572. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Lee, Y.S. Molecular functions of long noncoding transcripts in plants. J. Plant Biol. 2015, 58, 361–365. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Zhu, C.; Lu, G.; Guo, Y.; Zhou, Y.; Zhang, Z.; Zhao, Y.; Li, W.; Lu, Y.; Tang, W.; et al. Strand-specific RNA-seq reveals widespread occurrence of novel cis-natural antisense transcripts in rice. BMC Genom. 2012, 13, 721. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.H.; Liu, M.; Downie, B.; Liang, C.; Ji, G.; Li, Q.Q.; Hunt, A.G. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc. Natl. Acad. Sci. USA 2011, 108, 12533–12538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Deng, W.; Fan, X.; Liu, T.T.; He, G.; Chen, R.; Terzaghi, W.; Zhu, D.; Deng, X.W. Genomic Features and Regulatory Roles of Intermediate-Sized Non-Coding RNAs in Arabidopsis. Mol. Plant 2014, 7, 514–527. [Google Scholar] [CrossRef] [PubMed]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Bardou, F.; Ariel, F.; Simpson, C.G.; Romero-Barrios, N.; Laporte, P.; Balzergue, S.; Brown, J.W.S.; Crespi, M. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis. Dev. Cell 2014, 30, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.X.; Yan, B.X.; Qu, Y.Y.; Qin, F.F.; Yang, Y.T.; Hao, X.J.; Yu, J.J.; Zhao, Q.; Zhu, D.Y.; Ao, G.M. Zm401, a short-open reading-frame mRNA or noncoding RNA, is essential for tapetum and microspore development and can regulate the floret formation in maize. J. Cell Biochem. 2008, 105, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, H.J.; Fang, J.; Chu, C.C.; Wang, X.J. A long noncoding RNA involved in rice reproductive development by negatively regulating osa-miR160. Sci. Bull. 2017, 62, 470–475. [Google Scholar] [CrossRef]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Nonomura, K.I. Isolation and bioinformatic analyses of small RNAs interacting with germ cell-specific Argonaute in rice. In PIWI-Interacting RNAs. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2014; Volume 1093, pp. 1–249. [Google Scholar] [CrossRef]
- Ou, L.J.; Liu, Z.B.; Zhang, Z.Q.; Wei, G.; Zhang, Y.P.; Kang, L.Y.; Yang, B.Z.; Yang, S.; Lv, J.H.; Liu, Y.H. Noncoding and coding transcriptome analysis reveals the regulation roles of long noncoding RNAs in fruit development of hot pepper (Capsicum annuum L.). Plant Growth Regul. 2017, 83, 141–156. [Google Scholar] [CrossRef]
- Dietrich, A.; Wallet, C.; Iqbal, R.K.; Gualberto, J.M.; Lotfi, F. Organellar non-coding RNAs: Emerging regulation mechanisms. Biochimie 2015, 117, 48–62. [Google Scholar] [CrossRef] [PubMed]
- Rurek, M. Participation of non-coding RNAs in plant organelle biogenesis. Acta Biochim. Pol. 2016, 63, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.Q.; Stone, J.D.; Štorchová, H.; Sloan, D.B. High transcript abundance, RNA editing, and small RNAs originating from intergenic regions in the massive mitochondrial genome of the angiosperm Silene noctiflora. BMC Genom. 2015, 16, 938. [Google Scholar] [CrossRef] [PubMed]
- Richly, E.; Leister, D. NUMTs in sequenced eukaryotic genomes. Mol. Biol. Evol. 2004, 21, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Ruwe, H.; Wang, G.W.; Gusewski, S.; Schmitz-Linneweber, C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms. Nucleic Acids Res. 2016, 44, 7406–7417. [Google Scholar] [CrossRef] [PubMed]
- Holec, S.; Lange, H.; Kuhn, K.; Alioua, M.; Borner, T.; Gagliardi, D. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 2006, 26, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.D.; Koloušková, P.; Sloan, D.B.; Štorchová, H. Non-coding RNA may be associated with cytoplasmic male sterility in Silene vulgaris. J. Exp. Bot. 2017, 68, 1599–1612. [Google Scholar] [CrossRef] [PubMed]
| miRNA | Putative Target Genes | Target Gene Functions | References |
|---|---|---|---|
| Maize CMS C48-2 | |||
| Zma-miR397c | Laccase | Oxidation of phenolic substrates | |
| Zma-miR601 | Flavin-containing monooxygenase (FMO) | Auxin biosynthesis | [61] |
| Enoyl-CoA hydratase | Catabolism of fatty acids | ||
| Zma-miR604 | Monosaccharide transport protein 2 (STP2) | Uptake of glucose from callose degradation | |
| Brassica juncea hybrid | |||
| miR156a | SPL transcription factors | Floral transition, tapetum development | [57] |
| miR167a | Auxin response factor (ARF6/ARF8) | Anther dehiscence | |
| miR319a | TCP transcription factors | Floral induction | |
| miR395a | ATP sulphurylase (APS) | Sulphur metabolism | |
| Rice MeixiangA | |||
| osa-miR528-3p | F-box containing protein | Proteolytic turnover through proteasome | [42] |
| osa-miR1432-5p | Metal cation transporter | Cation homeostasis | |
| osa-miR2118c | NBS-LRR | Disease-resistance related proteins | |
| Brassica oleracea Bo01-12A | |||
| bol-miR157a | SPL transcription factors | Floral transition, tapetum development | [40] |
| bol-miR171a | SCARECROW-like (SCL) transcription factor | GA mediated action | |
| bol-miR172 | APETALA2 (AP2) transcription factor | Floral transition | |
| bol-miR824 | MADS-box transcription factor-like | Plant development | |
| Brassica rapa CMS-Ogura | |||
| bra-miR157a | SPL transcription factors | Floral transition, tapetum development | [59] |
| bra-miR158-3p | PPR-RFL | RNA metabolism in organelles | |
| bra-miR159a | MYB81 transcription factor | Flowering | |
| bra-miR164a | CUP SHAPED COTYLEDON 1 | Meristem development | |
| bra-miR172a | APETALA2 (AP2) transcription factor | Floral transition | |
| bra-miR5712 | VACUOLAR ATP SYNTHASE SUBUNIT A | Male gametophyte development | |
| bra-miR5716 | Zinc finger transcription factor | Drought stress response | |
| bra-miR6030 | CC-NBS-LRR | Disease-resistance related proteins | |
| Glycine max NJCMS1A | |||
| gma-miR166a-3p | HD-ZIPIII transcription factor | Vascular nad cell wall development | [60] |
| gma-miR169b | Nuclear factor Y (NF-YA) transcription factor | Flowering | |
| gma-miR171a | SCARECROW-like (SCL) transcription factor | GA mediated action | |
| gma-miR394b-5p | F-box protein | Proteolytic turnover through proteasome | |
| gma-miR395c | Sulphate transporter 2.1-like | Sulphur metabolism | |
| gma-miR396k-5p | bHLH79 transcription factor | Floral development | |
| gma-miR397a | Laccase | Oxidation of phenolic substrates | |
| gma-miR408c-3p | Plastocyanin-like | Copper metabolism | |
| Raphanus sativus CMS-WA | |||
| miR-158b-3p | PPR-RFL | RNA metabolism in organelles | [43] |
| miR161 | Mechanosensitive channel of small conductance-like 10 (MSL10) | Mechanosensitive ion channel, cell death induction | |
| miR395a | putative F-box/kelch-repeat (KFB) | Proteolytic turnover through proteasome | |
| Pummelo cybrid line | |||
| cga-miR156a.1 | SPL transcription factors | Floral transition, tapetum development | [58] |
| cga-miR399a.1 | UBC (ubiquitin-conjugating E2 enzyme) | Phosphate (Pi) homeostasis | |
| cga-miR827 | Basic leucine zipper (bZIP) | Pollen and flower development | |
© 2017 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Štorchová, H. The Role of Non-Coding RNAs in Cytoplasmic Male Sterility in Flowering Plants. Int. J. Mol. Sci. 2017, 18, 2429. https://doi.org/10.3390/ijms18112429
Štorchová H. The Role of Non-Coding RNAs in Cytoplasmic Male Sterility in Flowering Plants. International Journal of Molecular Sciences. 2017; 18(11):2429. https://doi.org/10.3390/ijms18112429
Chicago/Turabian StyleŠtorchová, Helena. 2017. "The Role of Non-Coding RNAs in Cytoplasmic Male Sterility in Flowering Plants" International Journal of Molecular Sciences 18, no. 11: 2429. https://doi.org/10.3390/ijms18112429




