Emerging Therapeutics to Overcome Chemoresistance in Epithelial Ovarian Cancer: A Mini-Review
Abstract
:1. Introduction
2. Chemoresistance
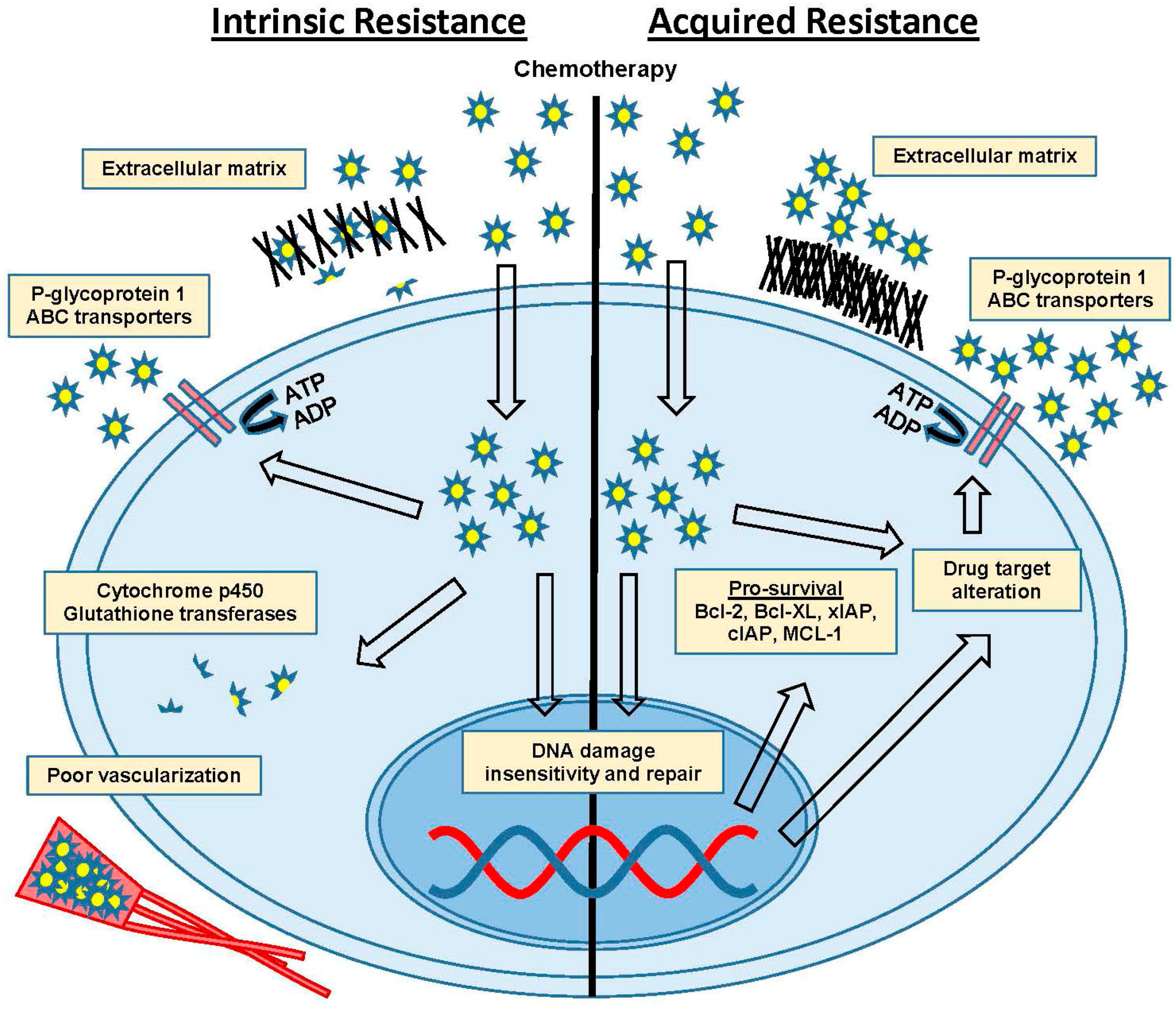
3. Intrinsic Versus Acquired Resistance
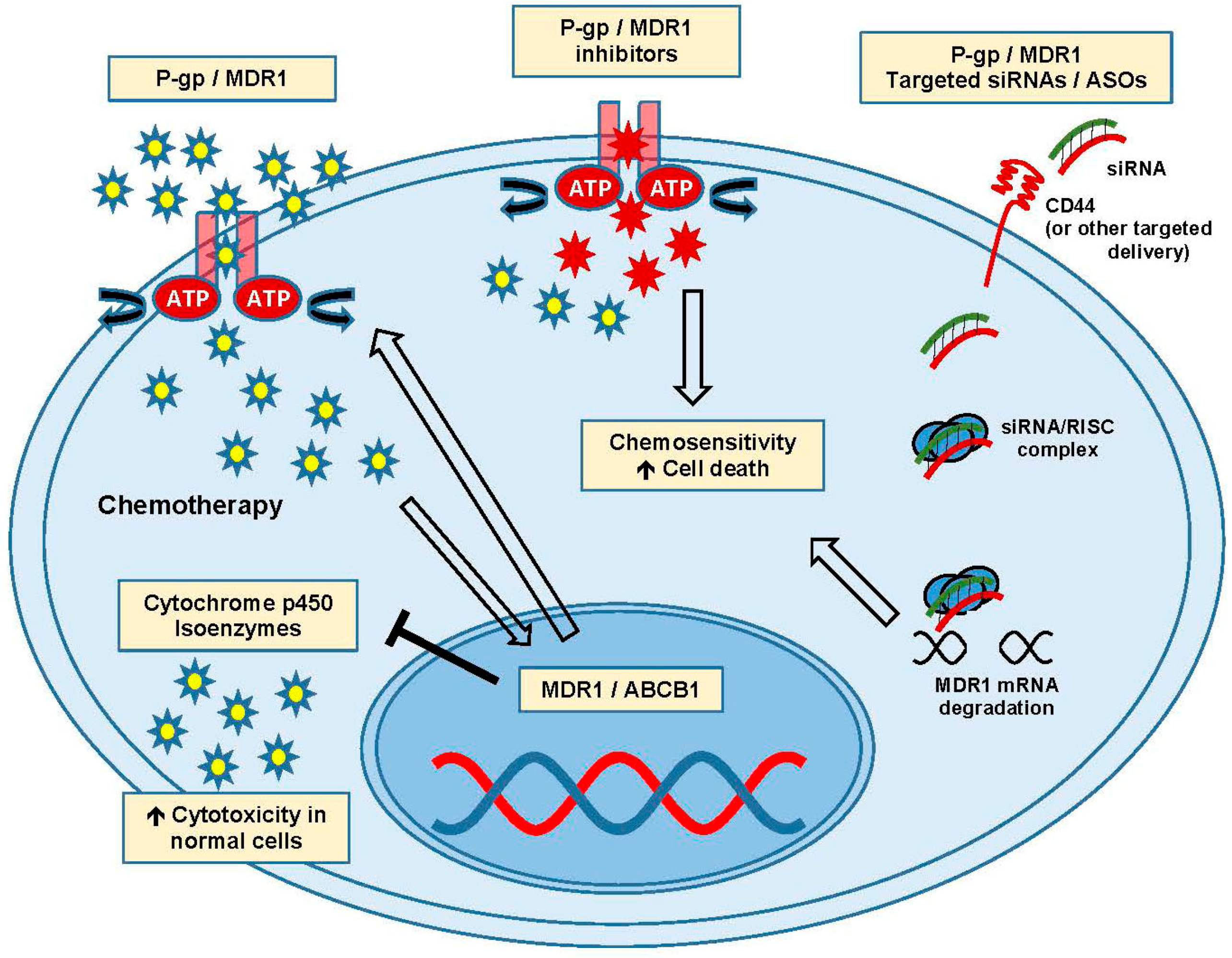
4. Efflux Pumps and MDR1
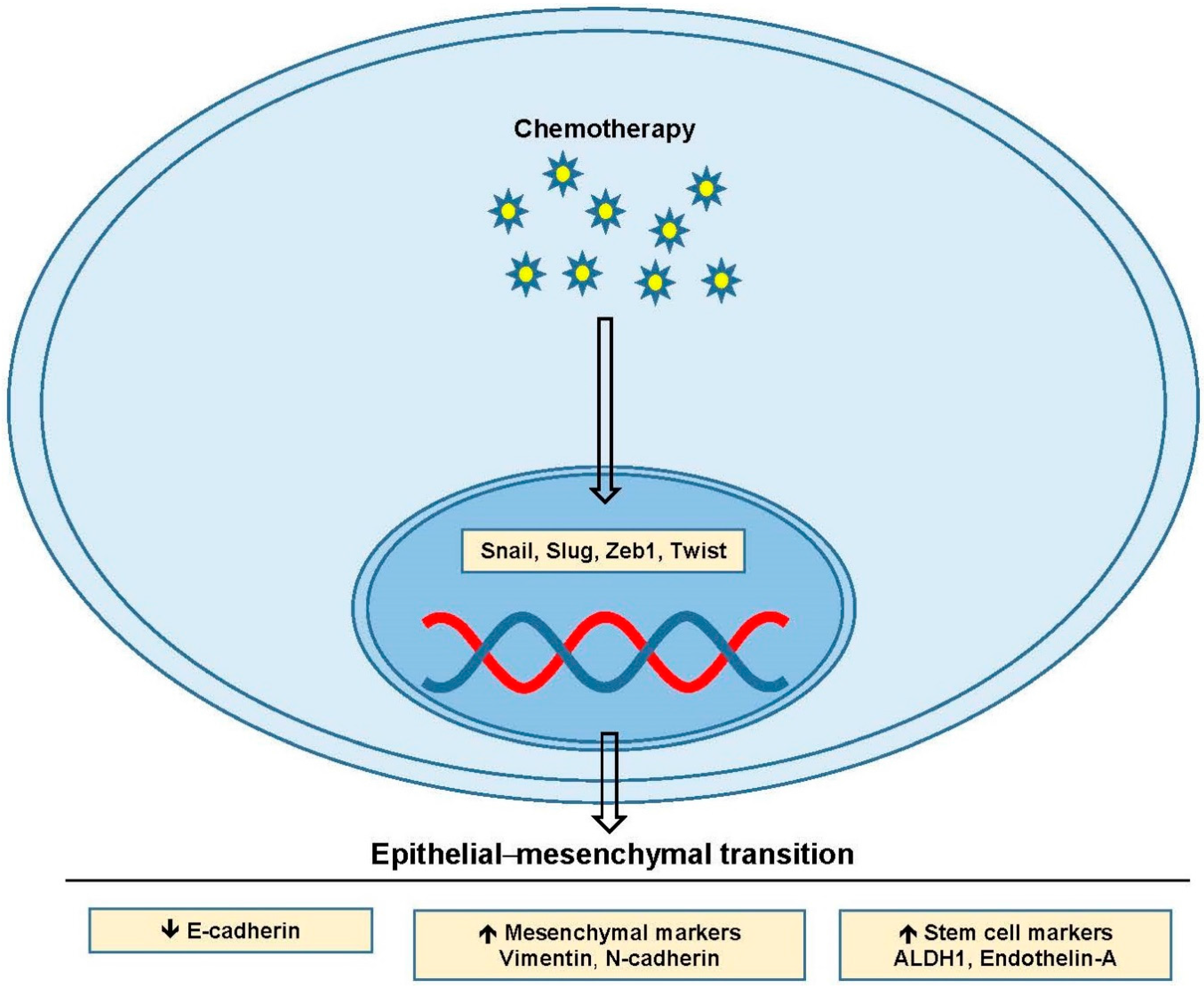
5. Epithelial–Mesenchymal Transition
6. DNA Damage Tolerance and Repair Capacity
7. Angiogenesis Inhibitors
8. PARP Inhibitors
9. Exosomes and Cancer
10. Checkpoint Inhibitors
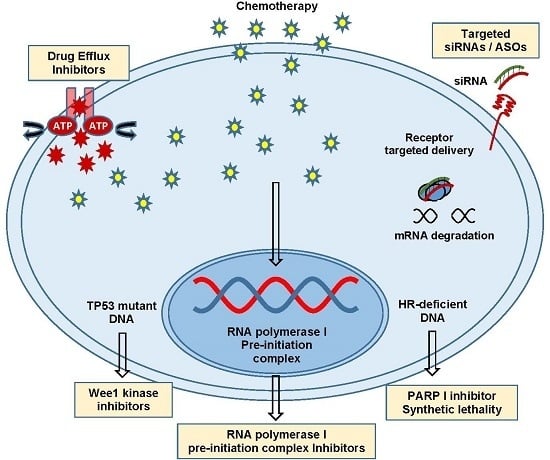
11. Ribosomal Biogenesis
12. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.-J.; Bast, R.C.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Hennessy, B.; Mills, G.B.; Mills, G.B. The biology of ovarian cancer: New opportunities for translation. Nat. Rev. Cancer 2009, 9, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, E. Ovarian cancer development and metastasis. Am. J. Pathol. 2010, 177, 1053–1064. [Google Scholar] [CrossRef] [PubMed]
- Harries, M.; Kaye, S.B. Recent advances in the treatment of epithelial ovarian cancer. Expert Opin. Investig. Drugs 2001, 10, 1715–1724. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Kaye, S.B. Ovarian cancer: Strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer 2003, 3, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Y.; Zhang, P.-Y. Recent perspectives of epithelial ovarian carcinoma (Review). Oncol. Lett. 2016, 12, 3055–3058. [Google Scholar] [CrossRef] [PubMed]
- Erickson, B.K.; Conner, M.G.; Landen, C.N. The role of the fallopian tube in the origin of ovarian cancer. Am. J. Obstet. Gynecol. 2013, 209, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Domcke, S.; Sinha, R.; Levine, D.A.; Sander, C.; Schultz, N. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat. Commun. 2013, 4, 2126. [Google Scholar] [CrossRef] [PubMed]
- Cannistra, S.A. Cancer of the Ovary. N. Engl. J. Med. 2004, 35124351, 2519–2529. [Google Scholar] [CrossRef] [PubMed]
- Abubaker, K.; Latifi, A.; Luwor, R.; Nazaretian, S.; Zhu, H.; Quinn, M.A.; Thompson, E.W.; Findlay, J.K.; Ahmed, N. Short-term single treatment of chemotherapy results in the enrichment of ovarian cancer stem cell-like cells leading to an increased tumor burden. Mol. Cancer 2013, 12, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coward, J.I.; Middleton, K.; Murphy, F. New perspectives on targeted therapy in ovarian cancer. Int. J. Women’s Health 2015, 7, 189–203. [Google Scholar] [CrossRef] [PubMed]
- Bagnoli, M.; Granata, A.; Nicoletti, R.; Krishnamachary, B.; Bhujwalla, Z.M.; Canese, R.; Podo, F.; Canevari, S.; Iorio, E.; Mezzanzanica, D. Choline Metabolism Alteration: A Focus on Ovarian Cancer. Front. Oncol. 2016, 6, 153. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, L.N.; Chow, E.K.-H. Mechanisms of chemoresistance in cancer stem cells. Clin. Transl. Med. 2013, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Liu, X.; Chan, D.; Ngan, H. Mechanisms of Chemoresistance in Human Ovarian Cancer at a Glance. Gynecol. Obstet. 2012, 2, 1000e104. [Google Scholar] [CrossRef]
- Krzystyniak, J.; Ceppi, L.; Dizon, D.S.; Birrer, M.J. Epithelial ovarian cancer: The molecular genetics of epithelial ovarian cancer. Ann. Oncol. 2016, 27, i4–i10. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, Z.C.; Katre, A.A.; Steg, A.D.; Erickson, B.K.; Shah, M.M.; Alvarez, R.D.; Conner, M.G.; Schneider, D.; Chen, D.; Landen, C.N. Using heterogeneity of the patient-derived xenograft model to identify the chemoresistant population in ovarian cancer. Oncotarget 2014, 5, 8750–8764. [Google Scholar] [CrossRef] [PubMed]
- Frei, E.; Karon, M.; Levin, R.H.; Freireich, E.J.; Taylor, R.J.; Hananian, J.; Selawry, O.; Holland, J.F.; Hoogstraten, B.; Wolman, I.J.; et al. The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia. Blood 1965, 26, 642–656. [Google Scholar] [PubMed]
- Zahreddine, H.; Borden, K.L.B. Mechanisms and insights into drug resistance in cancer. Front. Pharmacol. 2013, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.-S.; Chen, G.-D.; Tsai, H.-J.; Lee, M.-S.; Wang, P.-H.; Liu, F.-S. Mechanisms Involved in Chemoresistance in Ovarian Cancer. Taiwan J. Obstet. Gynecol. 2005, 44, 209–217. [Google Scholar] [CrossRef]
- Tapia, G.; Diaz-padilla, I. Molecular Mechanisms of Platinum Resistance in Ovarian Cancer. Ovarian Cancer A Clin. Transl. Update 2013, 371. [Google Scholar] [CrossRef]
- Cooke, S.L.; Brenton, J.D. Evolution of platinum resistance in high-grade serous ovarian cancer. Lancet Oncol. 2011, 12, 1169–1174. [Google Scholar] [CrossRef]
- Salomon-Perzyński, A.; Salomon-Perzyńska, M.; Michalski, B.; Skrzypulec-Plinta, V. High-grade serous ovarian cancer: The clone wars. Arch. Gynecol. Obstet. 2017, 295, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Pastan, I.H. The Role of Multidrug Resistance Efflux Pumps in Cancer: Revisiting a JNCI Publication Exploring Expression of the MDR1 (P-glycoprotein) Gene. J. Natl. Cancer Inst. 2015, 107, djv222. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Antona, C.; Ingelman-Sundberg, M. Cytochrome P450 pharmacogenetics and cancer. Oncogene 2006, 25, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Sueyoshi, T.; Negishi, M. Phenobarbital response elements of cytochrome P450 genes and nuclear receptors. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Saggar, J.K.; Yu, M.; Tan, Q.; Tannock, I.F. The tumor microenvironment and strategies to improve drug distribution. Front. Oncol. 2013, 3, 154. [Google Scholar] [CrossRef] [PubMed]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumours. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Meads, M.B.; Gatenby, R.A.; Dalton, W.S. Environment-mediated drug resistance: A major contributor to minimal residual disease. Nat. Rev. Cancer 2009, 9, 665–674. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Lu, A.Y. Interindividual variability in inhibition and induction of cytochrome P450 enzymes. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 535. [Google Scholar] [CrossRef] [PubMed]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Carden, C.P.; Kaye, S.B. Beyond chemotherapy: Targeted therapies in ovarian cancer. Nat. Rev. Cancer 2009, 9, 167–181. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Hu, D.; Qiu, J.; Xie, X.; Ye, F.; Lu, W.-G. Overexpression of glycogen synthase kinase-3 in ovarian carcinoma cells with acquired paclitaxel resistance. Int. J. Gynecol. Cancer 2011, 21, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Patch, A.-M.; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; Bailey, P.J.; et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Groenendijk, F.H.; Bernards, R. Drug resistance to targeted therapies: Déjà vu all over again. Mol. Oncol. 2014, 8, 1067–1083. [Google Scholar] [CrossRef] [PubMed]
- Orr, G.A.; Verdier-Pinard, P.; McDaid, H.; Horwitz, S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene 2003, 22, 7280–7295. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A.; Charlton, P. Molecular chess? Hallmarks of anti-cancer drug resistance. BMC Cancer 2017, 17, 10. [Google Scholar] [CrossRef] [PubMed]
- Kavallaris, M.; Kuo, D.Y.; Burkhart, C.A.; Regl, D.L.; Norris, M.D.; Haber, M.; Horwitz, S.B. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J. Clin. Investig. 1997, 100, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.D.; Welsbie, D.S.; Tran, C.; Baek, S.H.; Chen, R.; Vessella, R.; Rosenfeld, M.G.; Sawyers, C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2004, 10, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Lee, K. Targeting multidrug resistance with small molecules for cancer therapy. Biomol. Ther. 2010, 18, 375–385. [Google Scholar] [CrossRef]
- Vaidyanathan, A.; Sawers, L.; Gannon, A.-L.; Chakravarty, P.; Scott, A.L.; Bray, S.E.; Ferguson, M.J.; Smith, G. ABCB1 (MDR1) induction defines a common resistance mechanism in paclitaxel- and olaparib-resistant ovarian cancer cells. Br. J. Cancer 2016, 115, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Hedditch, E.L.; Gao, B.; Russell, A.J.; Lu, Y.; Emmanuel, C.; Beesley, J.; Johnatty, S.E.; Chen, X.; Harnett, P.; George, J.; et al. ABCA transporter gene expression and poor outcome in epithelial ovarian cancer. J. Natl. Cancer Inst. 2014, 106. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, A.H. P-glycoprotein, a gatekeeper in the blood-brain barrier. Adv. Drug Deliv. Rev. 1999, 36, 179–194. [Google Scholar] [CrossRef]
- Johnatty, S.E.; Beesley, J.; Paul, J.; Fereday, S.; Spurdle, A.B.; Webb, P.M.; Byth, K.; Marsh, S.; McLeod, H.; Harnett, P.R.; et al. ABCB1 (MDR 1) Polymorphisms and Progression-Free Survival among Women with Ovarian Cancer following Paclitaxel/Carboplatin Chemotherapy. Clin. Cancer Res. 2008, 14, 5594–5601. [Google Scholar] [CrossRef] [PubMed]
- Johnatty, S.E.; Beesley, J.; Gao, B.; Chen, X.; Lu, Y.; Law, M.H.; Henderson, M.J.; Russell, A.J.; Hedditch, E.L.; Emmanuel, C.; et al. ABCB1 (MDR1) polymorphisms and ovarian cancer progression and survival: A comprehensive analysis from the Ovarian Cancer Association Consortium and The Cancer Genome Atlas. Gynecol. Oncol. 2013, 131, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Iyer, A.K.; Singh, A.; Choy, E.; Hornicek, F.J.; Amiji, M.M.; Duan, Z. MDR1 siRNA loaded hyaluronic acid-based CD44 targeted nanoparticle systems circumvent paclitaxel resistance in ovarian cancer. Sci. Rep. 2015, 5, 8509. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Li, X.; Meng, X.-N.; Yan, J.; Zong, Z. MicroRNA-873 mediates multidrug resistance in ovarian cancer cells by targeting ABCB1. Tumor Biol. 2016, 37, 10499–10506. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, S.; Meng, X.; Shang, H.; Guan, Y. Inhibition of mdr1 by G-quadruplex oligonucleotides and reversal of paclitaxel resistance in human ovarian cancer cells. Tumor Biol. 2015, 36, 6433–6443. [Google Scholar] [CrossRef] [PubMed]
- Planting, A.S.T.; Sonneveld, P.; van der Gaast, A.; Sparreboom, A.; van der Burg, M.E.L.; Luyten, G.P.M.; de Leeuw, K.; de Boer-Dennert, M.; Wissel, P.S.; Jewell, R.C.; et al. A phase I and pharmacologic study of the MDR converter GF120918 in combination with doxorubicin in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2005, 55, 91–99. [Google Scholar] [CrossRef] [PubMed]
- El-Awady, R.; Saleh, E.; Hashim, A.; Soliman, N.; Dallah, A.; Elrasheed, A.; Elakraa, G. The Role of Eukaryotic and Prokaryotic ABC Transporter Family in Failure of Chemotherapy. Front. Pharmacol. 2016, 7, 535. [Google Scholar] [CrossRef] [PubMed]
- Wandel, C.; Kim, R.B.; Kajiji, S.; Guengerich, P.; Wilkinson, G.R.; Wood, A.J. P-glycoprotein and cytochrome P-450 3A inhibition: Dissociation of inhibitory potencies. Cancer Res. 1999, 59, 3944–3948. [Google Scholar] [PubMed]
- Seiden, M.V.; Swenerton, K.D.; Matulonis, U.; Campos, S.; Rose, P.; Batist, G.; Ette, E.; Garg, V.; Fuller, A.; Harding, M.W.; et al. A phase II study of the MDR inhibitor biricodar (INCEL, VX-710) and paclitaxel in women with advanced ovarian cancer refractory to paclitaxel therapy. Gynecol. Oncol. 2002, 86, 302–310. [Google Scholar] [CrossRef] [PubMed]
- Kapse-Mistry, S.; Govender, T.; Srivastava, R.; Yergeri, M. Nanodrug delivery in reversing multidrug resistance in cancer cells. Front. Pharmacol. 2014, 5, 159. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Ohnuma, S.; Ambudkar, S.V. Improving cancer chemotherapy with modulators of ABC drug transporters. Curr. Drug Targets 2011, 12, 621–630. [Google Scholar] [CrossRef] [PubMed]
- Binkhathlan, Z.; Lavasanifar, A. P-glycoprotein inhibition as a therapeutic approach for overcoming multidrug resistance in cancer: Current status and future perspectives. Curr. Cancer Drug Targets 2013, 13, 326–346. [Google Scholar] [CrossRef] [PubMed]
- Weidner, L.D.; Fung, K.L.; Kannan, P.; Moen, J.K.; Kumar, J.S.; Mulder, J.; Innis, R.B.; Gottesman, M.M.; Hall, M.D. Tariquidar Is an Inhibitor and Not a Substrate of Human and Mouse P-glycoprotein. Drug Metab. Dispos. 2016, 44, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.-J.; Liang, Y.-J.; Huang, H.-B.; Zhao, H.-Y.; Wu, C.-P.; Wang, F.; Tao, L.-Y.; Zhang, C.-Z.; Dai, C.-L.; Tiwari, A.K.; et al. Apatinib (YN968D1) reverses multidrug resistance by inhibiting the efflux function of multiple ATP-binding cassette transporters. Cancer Res. 2010, 70, 7981–7991. [Google Scholar] [CrossRef] [PubMed]
- Chung, F.S.; Santiago, J.S.; De Jesus, M.F.M.; Trinidad, C.V.; See, M.F.E. Disrupting P-glycoprotein function in clinical settings: What can we learn from the fundamental aspects of this transporter? Am. J. Cancer Res. 2016, 6, 1583–1598. [Google Scholar] [PubMed]
- Zhang, Y.; Sriraman, S.K.; Kenny, H.A.; Luther, E.; Torchilin, V.; Lengyel, E. Reversal of Chemoresistance in Ovarian Cancer by Co-Delivery of a P-glycoprotein Inhibitor and Paclitaxel in a Liposomal Platform. Mol. Cancer Ther. 2016, 15, 2282–2293. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Iyer, A.K.; Singh, A.; Milane, L.; Choy, E.; Hornicek, F.J.; Amiji, M.M.; Duan, Z. Cluster of Differentiation 44 Targeted Hyaluronic Acid Based Nanoparticles for MDR1 siRNA Delivery to Overcome Drug Resistance in Ovarian Cancer. Pharm. Res. 2015, 32, 2097–2109. [Google Scholar] [CrossRef] [PubMed]
- Rivera, E.; Gomez, H. Chemotherapy resistance in metastatic breast cancer: The evolving role of ixabepilone. Breast Cancer Res. 2010, 12 (Suppl. 2), S2. [Google Scholar] [CrossRef] [PubMed]
- Vishnu, P.; Colon-Otero, G.; Kennedy, G.T.; Marlow, L.A.; Kennedy, W.P.; Wu, K.J.; Santoso, J.T.; Copland, J.A. RhoB mediates antitumor synergy of combined ixabepilone and sunitinib in human ovarian serous cancer. Gynecol. Oncol. 2012, 124, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Padilla, I.; Oza, A.M. Epothilones in the treatment of ovarian cancer. Future Oncol. 2011, 7, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Zagouri, F.; Sergentanis, T.; Chrysikos, D.; Bamias, A.; Dimopoulos, A.-M. Epothilones in epithelial ovarian, fallopian tube, or primary peritoneal cancer: A systematic review. Oncol. Targets Ther. 2015, 8, 2187. [Google Scholar] [CrossRef] [PubMed]
- Roque, D.M.; Ratner, E.S.; Silasi, D.-A.; Azodi, M.; Rutherford, T.J.; Schwartz, P.E.; Nelson, W.K.; Santin, A.D. Weekly ixabepilone with or without biweekly bevacizumab in the treatment of recurrent or persistent uterine and ovarian/primary peritoneal/fallopian tube cancers: A retrospective review. Gynecol. Oncol. 2015, 137, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Tropé, C.G.; Reich, R. Epithelial-Mesenchymal Transition in Ovarian Carcinoma. Front. Oncol. 2012, 2, 33. [Google Scholar] [CrossRef] [PubMed]
- Rosanò, L.; Cianfrocca, R.; Spinella, F.; Di Castro, V.; Nicotra, M.R.; Lucidi, A.; Ferrandina, G.; Natali, P.G.; Bagnato, A. Acquisition of chemoresistance and EMT phenotype is linked with activation of the endothelin A receptor pathway in ovarian carcinoma cells. Clin. Cancer Res. 2011, 17, 2350–2360. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Young, M.-J.; Wu, Y.-H.; Chiu, W.-T.; Weng, T.-Y.; Huang, Y.-F.; Chou, C.-Y. All-trans retinoic acid downregulates ALDH1-mediated stemness and inhibits tumour formation in ovarian cancer cells. Carcinogenesis 2015, 36, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Bagnato, A.; Rosanò, L. Understanding and overcoming chemoresistance in ovarian cancer: Emerging role of the endothelin axis. Curr. Oncol. 2012, 19, 36–38. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Kobayashi, S.; Yamada, D.; Nagano, H.; Tomokuni, A.; Tomimaru, Y.; Noda, T.; Gotoh, K.; Asaoka, T.; Wada, H.; et al. A Histone Deacetylase Inhibitor Suppresses Epithelial-Mesenchymal Transition and Attenuates Chemoresistance in Biliary Tract Cancer. PLoS ONE 2016, 11, e0145985. [Google Scholar] [CrossRef] [PubMed]
- Kajiyama, H.; Shibata, K.; Terauchi, M.; Yamashita, M.; Ino, K.; Nawa, A.; Kikkawa, F. Chemoresistance to paclitaxel induces epithelial-mesenchymal transition and enhances metastatic potential for epithelial ovarian carcinoma cells. Int. J. Oncol. 2007, 31, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Haslehurst, A.M.; Koti, M.; Dharsee, M.; Nuin, P.; Evans, K.; Geraci, J.; Childs, T.; Chen, J.; Li, J.; Weberpals, J.; et al. EMT transcription factors snail and slug directly contribute to cisplatin resistance in ovarian cancer. BMC Cancer 2012, 12, 91. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Xu, Z.; El-Sehemy, A.; Steed, H.; Fu, Y. Notch3 induces epithelial-mesenchymal transition and attenuates carboplatin-induced apoptosis in ovarian cancer cells. Gynecol. Oncol. 2013, 130, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Latifi, A.; Abubaker, K.; Castrechini, N.; Ward, A.C.; Liongue, C.; Dobill, F.; Kumar, J.; Thompson, E.W.; Quinn, M.A.; Findlay, J.K.; et al. Cisplatin treatment of primary and metastatic epithelial ovarian carcinomas generates residual cells with mesenchymal stem cell-like profile. J. Cell. Biochem. 2011, 112, 2850–2864. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.P.; Dupre, T.V.; Siskind, L.J.; Beverly, L.J. Common cytotoxic chemotherapeutics induce epithelial-mesenchymal transition (EMT) downstream of ER stress. Oncotarget 2017, 8, 22625–22639. [Google Scholar] [CrossRef] [PubMed]
- Rosano, L.; Cianfrocca, R.; Tocci, P.; Spinella, F.; Di Castro, V.; Caprara, V.; Semprucci, E.; Ferrandina, G.; Natali, P.G.; Bagnato, A. Endothelin A Receptor/-Arrestin Signaling to the Wnt Pathway Renders Ovarian Cancer Cells Resistant to Chemotherapy. Cancer Res. 2014, 74, 7453–7464. [Google Scholar] [CrossRef] [PubMed]
- Miow, Q.H.; Tan, T.Z.; Ye, J.; Lau, J.A.; Yokomizo, T.; Thiery, J.; Mori, S. Epithelial-mesenchymal status renders differential responses to cisplatin in ovarian cancer. Oncogene 2015, 34, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Pradeep, S.; Kim, S.; Wu, S.; Nishimura, M.; Chaluvally-Raghavan, P.; Miyake, T.; Pecot, C.; Kim, S.J.; Choi, H.; Bischoff, F.; et al. Hematogenous Metastasis of Ovarian Cancer: Rethinking Mode of Spread. Cancer Cell 2014, 26, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Marchini, S.; Fruscio, R.; Clivio, L.; Beltrame, L.; Porcu, L.; Nerini, I.F.; Cavalieri, D.; Chiorino, G.; Cattoretti, G.; Mangioni, C.; et al. Resistance to platinum-based chemotherapy is associated with epithelial to mesenchymal transition in epithelial ovarian cancer. Eur. J. Cancer 2013, 49, 520–530. [Google Scholar] [CrossRef] [PubMed]
- Tomkinson, H.; Kemp, J.; Oliver, S.; Swaisland, H.; Taboada, M.; Morris, T. Pharmacokinetics and tolerability of zibotentan (ZD4054) in subjects with hepatic or renal impairment: Two open-label comparative studies. BMC Clin. Pharmacol. 2011, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.; Tho, L.M.; Xu, N.; Gillespie, D.A. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 2010, 108, 73–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Paull, T.T. ATM Activation by DNA Double-Strand Breaks through the Mre11-Rad50-Nbs1 Complex. Science 2005, 308, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Fattaey, A.; Booher, R.N. Myt1: A Wee1-type kinase that phosphorylates Cdc2 on residue Thr14. Prog. Cell Cycle Res. 1997, 3, 233–240. [Google Scholar] [PubMed]
- Pines, J.; Hunter, T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994, 13, 3772–3781. [Google Scholar] [PubMed]
- McGowan, C.H.; Russell, P. Human Wee1 kinase inhibits cell division by phosphorylating p34cdc2 exclusively on Tyr15. EMBO J. 1993, 12, 75–85. [Google Scholar] [PubMed]
- Wells, N.J.; Watanabe, N.; Tokusumi, T.; Jiang, W.; Verdecia, M.A.; Hunter, T. The C-terminal domain of the Cdc2 inhibitory kinase Myt1 interacts with Cdc2 complexes and is required for inhibition of G(2)/M progression. J. Cell Sci. 1999, 112, 3361–3371. [Google Scholar] [PubMed]
- Atherton-Fessler, S.; Liu, F.; Gabrielli, B.; Lee, M.S.; Peng, C.Y.; Piwnica-Worms, H. Cell cycle regulation of the p34cdc2 inhibitory kinases. Mol. Biol. Cell 1994, 5, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signaling. Cell 1995, 80, 225–236. [Google Scholar] [CrossRef]
- Ho, G.Y.; Woodward, N.; Coward, J.I.G. Cisplatin versus carboplatin: Comparative review of therapeutic management in solid malignancies. Crit. Rev. Oncol. Hematol. 2016, 102, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Lippard, S.J. Cellular processing of platinum anticancer drugs. Nat. Rev. Drug Discov. 2005, 4, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Nojima, K.; Hochegger, H.; Saberi, A.; Fukushima, T.; Kikuchi, K.; Yoshimura, M.; Orelli, B.J.; Bishop, D.K.; Hirano, S.; Ohzeki, M.; et al. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 2005, 65, 11704–11711. [Google Scholar] [CrossRef] [PubMed]
- Reinbolt, R.E.; Hays, J.L. The Role of PARP Inhibitors in the Treatment of Gynecologic Malignancies. Front. Oncol. 2013, 3, 237. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A. A Synthetic Lethal Therapeutic Approach: Poly(ADP) Ribose Polymerase Inhibitors for the Treatment of Cancers Deficient in DNA Double-Strand Break Repair. J. Clin. Oncol. 2008, 26, 3785–3790. [Google Scholar] [CrossRef] [PubMed]
- Bache, M.; Pigorsch, S.; Dunst, J.; Würl, P.; Meye, A.; Bartel, F.; Schmidt, H.; Rath, F.W.; Taubert, H. Loss of G2/M arrest correlates with radiosensitization in two human sarcoma cell lines with mutant p53. Int. J. Cancer 2001, 96, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Matheson, C.J.; Backos, D.S.; Reigan, P. Targeting WEE1 Kinase in Cancer. Trends Pharmacol. Sci. 2016, 37, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Venezia, T.A.; Merchant, A.A.; Ramos, C.A.; Whitehouse, N.L.; Young, A.S.; Shaw, C.A.; Goodell, M.A. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004, 2, e301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selvakumaran, M.; Pisarcik, D.A.; Bao, R.; Yeung, A.T.; Hamilton, T.C. Enhanced cisplatin cytotoxicity by disturbing the nucleotide excision repair pathway in ovarian cancer cell lines. Cancer Res. 2003, 63, 1311–1316. [Google Scholar] [PubMed]
- Leijen, S.; Beijnen, J.H.; Schellens, J.H.M. Abrogation of the G2 checkpoint by inhibition of Wee-1 kinase results in sensitization of p53-deficient tumor cells to DNA-damaging agents. Curr. Clin. Pharmacol. 2010, 5, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Osman, A.A.; Monroe, M.M.; Ortega Alves, M.V.; Patel, A.A.; Katsonis, P.; Fitzgerald, A.L.; Neskey, D.M.; Frederick, M.J.; Woo, S.H.; Caulin, C.; et al. Wee-1 Kinase Inhibition Overcomes Cisplatin Resistance Associated with High-Risk TP53 Mutations in Head and Neck Cancer through Mitotic Arrest Followed by Senescence. Mol. Cancer Ther. 2015, 14, 608–619. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, O.; Shinkawa, M.; Torimura, T.; Nakamura, T.; Selvendiran, K.; Sakamoto, M.; Koga, H.; Ueno, T.; Sata, M. Cell cycle regulation by the Wee1 Inhibitor PD0166285, Pyrido [2,3-d] pyimidine, in the B16 mouse melanoma cell line. BMC Cancer 2006, 6, 292. [Google Scholar] [CrossRef] [PubMed]
- PosthumaDeBoer, J.; Würdinger, T.; Graat, H.C.; van Beusechem, V.W.; Helder, M.N.; van Royen, B.J.; Kaspers, G.J. WEE1 inhibition sensitizes osteosarcoma to radiotherapy. BMC Cancer 2011, 11, 156. [Google Scholar] [CrossRef] [PubMed]
- Heijink, A.M.; Blomen, V.A.; Bisteau, X.; Degener, F.; Matsushita, F.Y.; Kaldis, P.; Foijer, F.; van Vugt, M.A.T.M. A haploid genetic screen identifies the G1/S regulatory machinery as a determinant of Wee1 inhibitor sensitivity. Proc. Natl. Acad. Sci. USA 2015, 112, 15160–15165. [Google Scholar] [CrossRef] [PubMed]
- Trimbos, J.B.; Parmar, M.; Vergote, I.; Guthrie, D.; Bolis, G.; Colombo, N.; Vermorken, J.B.; Torri, V.; Mangioni, C.; Pecorelli, S.; et al. International Collaborative Ovarian Neoplasm trial 1 and Adjuvant ChemoTherapy In Ovarian Neoplasm trial: Two parallel randomized phase III trials of adjuvant chemotherapy in patients with early-stage ovarian carcinoma. J. Natl. Cancer Inst. 2003, 95, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Reinthaller, A. Antiangiogenic therapies in ovarian cancer. Memo 2016, 9, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Han, E.S.; Monk, B.J. Bevacizumab in the treatment of ovarian cancer. Expert Rev. Anticancer Ther. 2007, 7, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Aghajanian, C. The role of bevacizumab in ovarian cancer—An evolving story. Gynecol. Oncol. 2006, 102, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Rigamonti, N.; Kadioglu, E.; Keklikoglou, I.; Wyser Rmili, C.; Leow, C.C.; De Palma, M. Role of Angiopoietin-2 in Adaptive Tumor Resistance to VEGF Signaling Blockade. Cell Rep. 2014, 8, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Brunckhorst, M.K.; Xu, Y.; Lu, R.; Yu, Q. Angiopoietins Promote Ovarian Cancer Progression by Establishing a Procancer Microenvironment. Am. J. Pathol. 2014, 184, 2285–2296. [Google Scholar] [CrossRef] [PubMed]
- Pujade-Lauraine, E.; Hilpert, F.; Weber, B.; Reuss, A.; Poveda, A.; Kristensen, G.; Sorio, R.; Vergote, I.; Witteveen, P.; Bamias, A.; et al. Bevacizumab Combined With Chemotherapy for Platinum-Resistant Recurrent Ovarian Cancer: The AURELIA Open-Label Randomized Phase III Trial. J. Clin. Oncol. 2014, 32, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Monk, B.J.; Poveda, A.; Vergote, I.; Raspagliesi, F.; Fujiwara, K.; Bae, D.-S.; Oaknin, A.; Ray-Coquard, I.; Provencher, D.M.; Karlan, B.Y.; et al. Anti-angiopoietin therapy with trebananib for recurrent ovarian cancer (TRINOVA-1): A randomised, multicentre, double-blind, placebo-controlled phase 3 trial. Lancet Oncol. 2014, 15, 799–808. [Google Scholar] [CrossRef]
- McLachlan, J.; George, A.; Banerjee, S. The current status of PARP inhibitors in ovarian cancer. Tumori J. 2016, 102, 433–440. [Google Scholar] [CrossRef] [PubMed]
- Javle, M.; Curtin, N.J. The role of PARP in DNA repair and its therapeutic exploitation. Br. J. Cancer 2011, 105, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Dockery, L.E.; Gunderson, C.C.; Moore, K.N. Rucaparib: The past, present, and future of a newly approved PARP inhibitor for ovarian cancer. Onco Targets Ther. 2017, 10, 3029–3037. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.G.; Sarkaria, J.N.; Kaufmann, S.H. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl. Acad. Sci. USA 2011, 108, 3406–3411. [Google Scholar] [CrossRef] [PubMed]
- Evans, T.; Matulonis, U. PARP inhibitors in ovarian cancer: Evidence, experience and clinical potential. Ther. Adv. Med. Oncol. 2017, 9, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, S.; Beaver, J.A.; Horton, S.; Fernandes, L.L.; Tang, S.; Horne, H.N.; Liu, J.; Liu, C.; Schrieber, S.J.; Yu, J.; et al. FDA Approval Summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin. Cancer Res. 2017. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Lin, K.K.; Oza, A.M.; Scott, C.L.; Giordano, H.; Sun, J.; Konecny, G.E.; Coleman, R.L.; Tinker, A.V.; O’Malley, D.M.; et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): An international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 75–87. [Google Scholar] [CrossRef]
- Kim, Y.; Kim, A.; Sharip, A.; Sharip, A.; Jiang, J.; Yang, Q.; Xie, Y. Reverse the Resistance to PARP Inhibitors. Int. J. Biol. Sci. 2017, 13, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A. Drug resistance caused by reversion mutation. Cancer Res. 2008, 68, 10021–10023. [Google Scholar] [CrossRef] [PubMed]
- Dhillon, K.K.; Taniguchi, T. Resistance to PARP Inhibitors Mediated by Secondary BRCA1/2 Mutations; Humana Press: Cham, Switzerland, 2015; pp. 431–452. [Google Scholar]
- Barber, L.J.; Sandhu, S.; Chen, L.; Campbell, J.; Kozarewa, I.; Fenwick, K.; Assiotis, I.; Rodrigues, D.N.; Reis-Filho, J.S.; Moreno, V.; et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J. Pathol. 2013, 229, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Neijenhuis, S.; Bajrami, I.; Miller, R.; Lord, C.J.; Ashworth, A. Identification of miRNA modulators to PARP inhibitor response. DNA Repair 2013, 12, 394–402. [Google Scholar] [CrossRef] [PubMed]
- Ter Brugge, P.; Kristel, P.; van der Burg, E.; Boon, U.; de Maaker, M.; Lips, E.; Mulder, L.; de Ruiter, J.; Moutinho, C.; Gevensleben, H.; et al. Mechanisms of Therapy Resistance in Patient-Derived Xenograft Models of BRCA1-Deficient Breast Cancer. J. Natl. Cancer Inst. 2016, 108, djw148. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.L.; Brough, R.; Lord, C.J.; Natrajan, R.; Vatcheva, R.; Levine, D.A.; Boyd, J.; Reis-Filho, J.S.; Ashworth, A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature 2008, 451, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Krais, J.J.; Bernhardy, A.J.; Nicolas, E.; Cai, K.Q.; Harrell, M.I.; Kim, H.H.; George, E.; Swisher, E.M.; Simpkins, F.; et al. RING domain-deficient BRCA1 promotes PARP inhibitor and platinum resistance. J. Clin. Investig. 2016, 126, 3145–3157. [Google Scholar] [CrossRef] [PubMed]
- Rottenberg, S.; Jaspers, J.E.; Kersbergen, A.; van der Burg, E.; Nygren, A.O.H.; Zander, S.A.L.; Derksen, P.W.B.; de Bruin, M.; Zevenhoven, J.; Lau, A.; et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. USA 2008, 105, 17079–17084. [Google Scholar] [CrossRef] [PubMed]
- Issaeva, N.; Thomas, H.D.; Djurenovic, T.; Jaspers, J.E.; Stoimenov, I.; Kyle, S.; Pedley, N.; Gottipati, P.; Zur, R.; Sleeth, K.; et al. 6-Thioguanine Selectively Kills BRCA2-Defective Tumors and Overcomes PARP Inhibitor Resistance. Cancer Res. 2010, 70, 6268–6276. [Google Scholar] [CrossRef] [PubMed]
- Trams, E.G.; Lauter, C.J.; Salem, N.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Rashed, M.H.; Bayraktar, E.; Helal, G.K.; Abd-Ellah, M.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef] [PubMed]
- Corrado, C.; Raimondo, S.; Chiesi, A.; Ciccia, F.; De Leo, G.; Alessandro, R. Exosomes as intercellular signaling organelles involved in health and disease: Basic science and clinical applications. Int. J. Mol. Sci. 2013, 14, 5338–5366. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X. The emerging roles and therapeutic potential of exosomes in epithelial ovarian cancer. Mol. Cancer 2017, 16, 92. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, Y.; Zhao, M. Exosome-Based Cancer Therapy: Implication for Targeting Cancer Stem Cells. Front. Pharmacol. 2016, 7, 533. [Google Scholar] [CrossRef] [PubMed]
- Bard, M.P.; Hegmans, J.P.; Hemmes, A.; Luider, T.M.; Willemsen, R.; Severijnen, L.-A.A.; van Meerbeeck, J.P.; Burgers, S.A.; Hoogsteden, H.C.; Lambrecht, B.N. Proteomic Analysis of Exosomes Isolated from Human Malignant Pleural Effusions. Am. J. Respir. Cell Mol. Biol. 2004, 31, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Bhatnagar, S. Exosome Function: From Tumor Immunology to Pathogen Biology. Traffic 2008, 9, 871–881. [Google Scholar] [CrossRef] [PubMed]
- Bach, D.-H.; Hong, J.-Y.; Park, H.J.; Lee, S.K. The role of exosomes and miRNAs in drug-resistance of cancer cells. Int. J. Cancer 2017, 141, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.; Atay, S.; Banskota, S.; Artale, B.; Schmitt, S.; Godwin, A.K. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget 2017, 8, 11917–11936. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.-L.; Zhuang, T.; Xing, B.-H.; Li, N.; Li, Q. Exosomal DNMT1 mediates cisplatin resistance in ovarian cancer. Cell Biochem. Funct. 2017, 35, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.-S.; Roh, T.-Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.-K.; Gho, Y.S. Bioinspired Exosome-Mimetic Nanovesicles for Targeted Delivery of Chemotherapeutics to Malignant Tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef] [PubMed]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Lang, J. Programmed death-1 pathway blockade produces a synergistic antitumor effect: Combined application in ovarian cancer. J. Gynecol. Oncol. 2017, 28, e64. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.-Y.; Francois, A.; McGray, A.R.; Miliotto, A.; Odunsi, K. Compensatory upregulation of PD-1, LAG-3, and CTLA-4 limits the efficacy of single-agent checkpoint blockade in metastatic ovarian cancer. Oncoimmunology 2017, 6, e1249561. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.; Huemer, F.; Mlineritsch, B.; Greil, R. Immune checkpoint blockade in ovarian cancer. Memo 2016, 9, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Hamanishi, J.; Mandai, M.; Ikeda, T.; Minami, M.; Kawaguchi, A.; Murayama, T.; Kanai, M.; Mori, Y.; Matsumoto, S.; Chikuma, S.; et al. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 2015, 33, 4015–4022. [Google Scholar] [CrossRef] [PubMed]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Aust, S.; Felix, S.; Auer, K.; Bachmayr-Heyda, A.; Kenner, L.; Dekan, S.; Meier, S.M.; Gerner, C.; Grimm, C.; Pils, D. Absence of PD-L1 on tumor cells is associated with reduced MHC I expression and PD-L1 expression increases in recurrent serous ovarian cancer. Sci. Rep. 2017, 7, 42929. [Google Scholar] [CrossRef] [PubMed]
- Derenzini, M.; Montanaro, L.; Treré, D. What the nucleolus says to a tumour pathologist. Histopathology 2009, 54, 753–762. [Google Scholar] [CrossRef] [PubMed]
- Sirri, V.; Urcuqui-Inchima, S.; Roussel, P.; Hernandez-Verdun, D. Nucleolus: The fascinating nuclear body. Histochem. Cell Biol. 2008, 129, 13–31. [Google Scholar] [CrossRef] [PubMed]
- Deisenroth, C.; Zhang, Y. The Ribosomal Protein-Mdm2-p53 Pathway and Energy Metabolism: Bridging the Gap between Feast and Famine. Genes Cancer 2011, 2, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Boulon, S.; Westman, B.J.; Hutten, S.; Boisvert, F.M.; Lamond, A.I. The Nucleolus under Stress. Mol. Cell 2010, 40, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Woods, S.J.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim. Biophys. Acta 2015, 1849, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Quin, J.E.; Devlin, J.R.; Cameron, D.; Hannan, K.M.; Pearson, R.B.; Hannan, R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, K.; Colis, L.; Liu, H.; Trivedi, R.; Moubarek, M.S.; Moore, H.M.; Bai, B.; Rudek, M.A.; Bieberich, C.J.; Laiho, M. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell 2014, 25, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Pickard, A.J.; Bierbach, U. The cell’s nucleolus: An emerging target for chemotherapeutic intervention. ChemMedChem 2013, 8, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Frank, D.; Son, J.; Hannan, K.; Hannan, R.; Chan, K.; Pearson, R.; Sanij, E. The Potential of Targeting Ribosome Biogenesis in High-Grade Serous Ovarian Cancer. Int. J. Mol. Sci. 2017, 18, 210. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.; Zomerdijk, J.C.B.M. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006, 203–216. [Google Scholar] [CrossRef]
- Ray, S.; Panova, T.; Miller, G.; Volkov, A.; Porter, A.C.G.; Russell, J.; Panov, K.I.; Zomerdijk, J.C.B.M. Topoisomerase IIα promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat. Commun. 2013, 4, 1598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poortinga, G.; Quinn, L.; Hannan, R. Targeting RNA polymerase I to treat MYC-driven cancer. Oncogene 2014, 34, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Haddach, M.; Schwaebe, M.K.; Michaux, J.; Nagasawa, J.; O’Brien, S.E.; Whitten, J.P.; Pierre, F.; Kerdoncuff, P.; Darjania, L.; Stansfield, R.; et al. Discovery of CX-5461, the first direct and selective inhibitor of RNA polymerase I, for cancer therapeutics. ACS Med. Chem. Lett. 2012, 3, 602–606. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Di Antonio, M.; Mckinney, S.; Mathew, V.; Neil, N.J.O.; Dos Santos, N.; Wei, V.; Garcia, J.; Yap, D.; Le, D.; et al. CX-5461 and CX-3543 are G-quadruplex stabilizers with selective synthetic lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Pang, S.; Zhang, W.; Guo, X.; Wang, J.; Zhang, Y.; Liu, Y.; Wu, X.; Jiang, F. Therapeutic Targeting of RNA Polymerase I With the Small-Molecule CX-5461 for Prevention of Arterial Injury-Induced Neointimal Hyperplasia. Arterioscler. Thromb. Vasc. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, K.; Colis, L.; Liu, H.; Jaamaa, S.; Zhang, Z.; Af Hallstrom, T.; Moore, H.M.; Sirajuddin, P.; Laiho, M. Small Molecule BMH-Compounds That Inhibit RNA Polymerase I and Cause Nucleolar Stress. Mol. Cancer Ther. 2014, 13, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Colis, L.; Peltonen, K.; Sirajuddin, P.; Liu, H.; Sanders, S.; Ernst, G.; Barrow, J.C.; Laiho, M. DNA intercalator BMH-21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget 2014, 5, 4361–4369. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cornelison, R.; Llaneza, D.C.; Landen, C.N. Emerging Therapeutics to Overcome Chemoresistance in Epithelial Ovarian Cancer: A Mini-Review. Int. J. Mol. Sci. 2017, 18, 2171. https://doi.org/10.3390/ijms18102171
Cornelison R, Llaneza DC, Landen CN. Emerging Therapeutics to Overcome Chemoresistance in Epithelial Ovarian Cancer: A Mini-Review. International Journal of Molecular Sciences. 2017; 18(10):2171. https://doi.org/10.3390/ijms18102171
Chicago/Turabian StyleCornelison, Robert, Danielle C. Llaneza, and Charles N. Landen. 2017. "Emerging Therapeutics to Overcome Chemoresistance in Epithelial Ovarian Cancer: A Mini-Review" International Journal of Molecular Sciences 18, no. 10: 2171. https://doi.org/10.3390/ijms18102171