Lipid and Glycolipid Isomer Analyses Using Ultra-High Resolution Ion Mobility Spectrometry Separations
Abstract
:1. Introduction
2. Results and Discussion
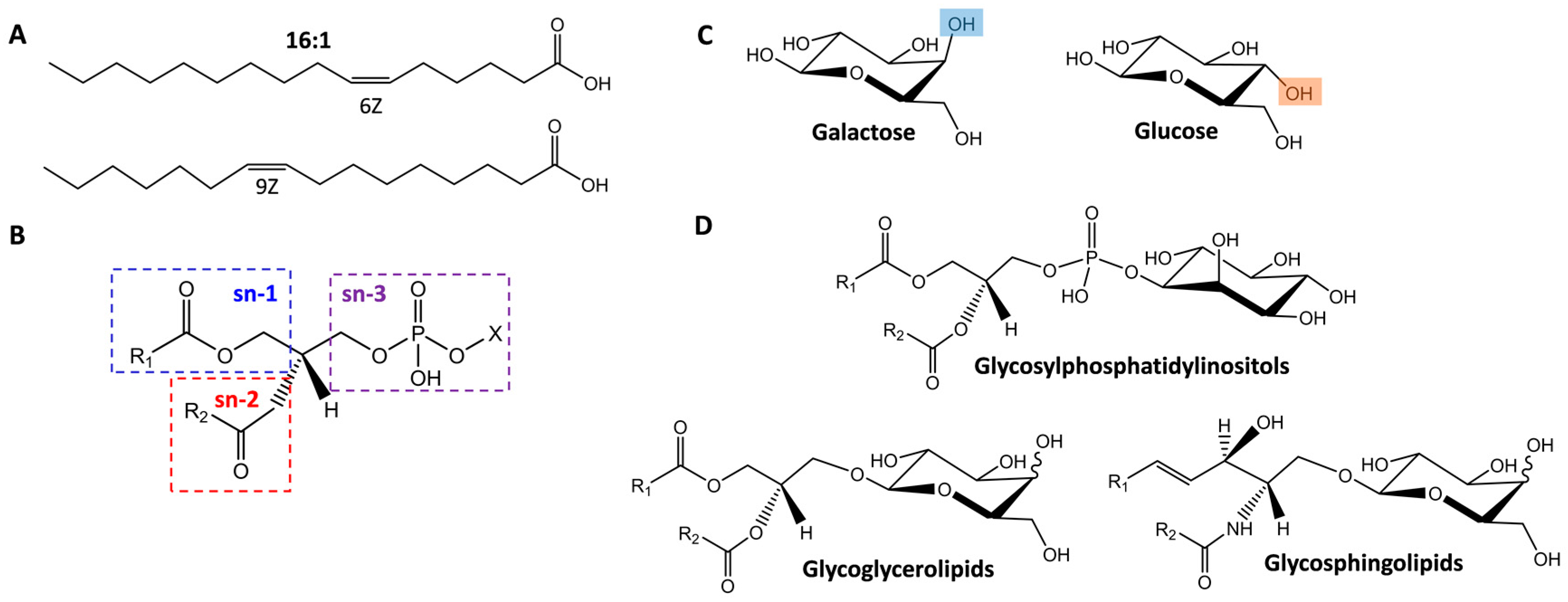
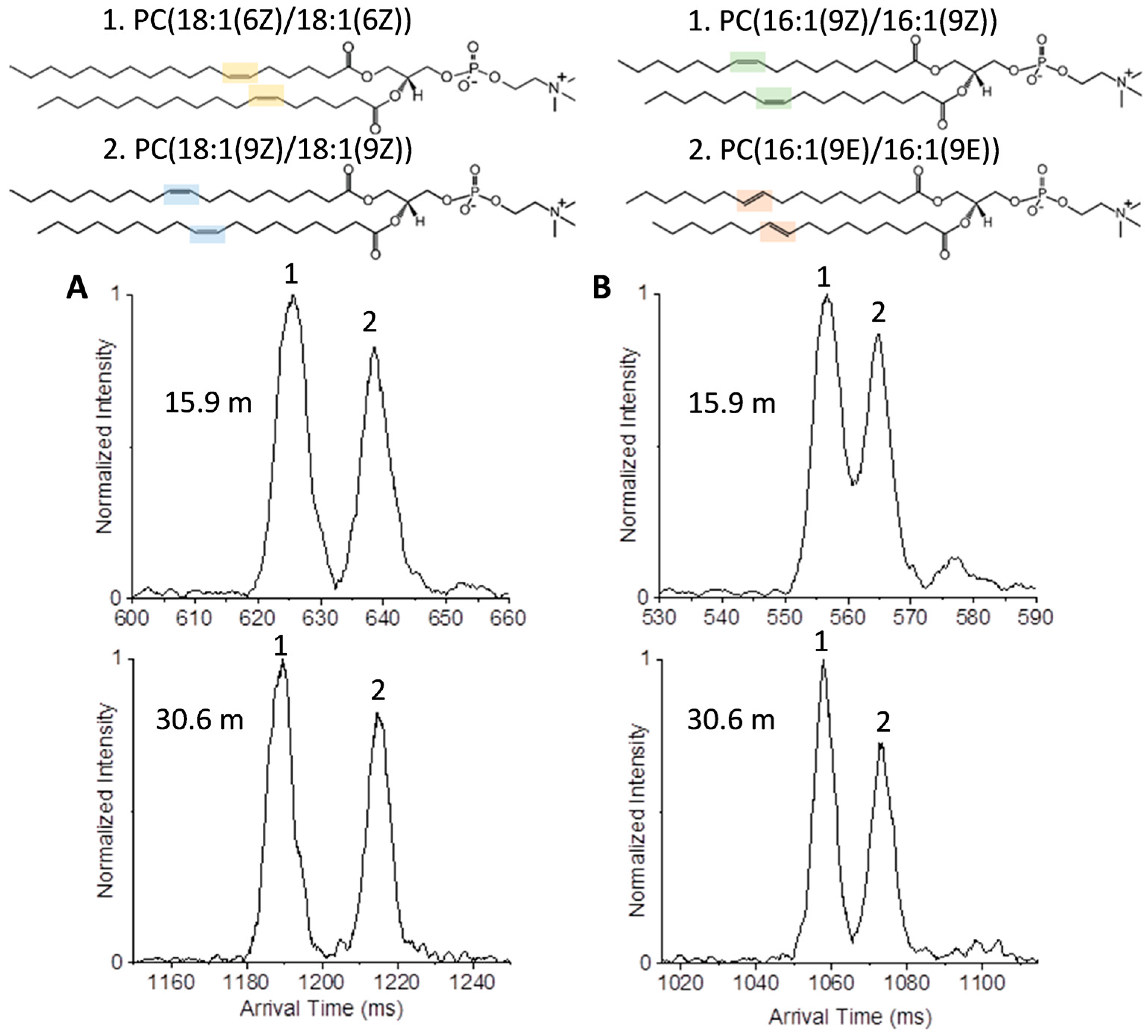
2.1. Isomer Separation of Double Bond Positions and Cis/Trans Orientations
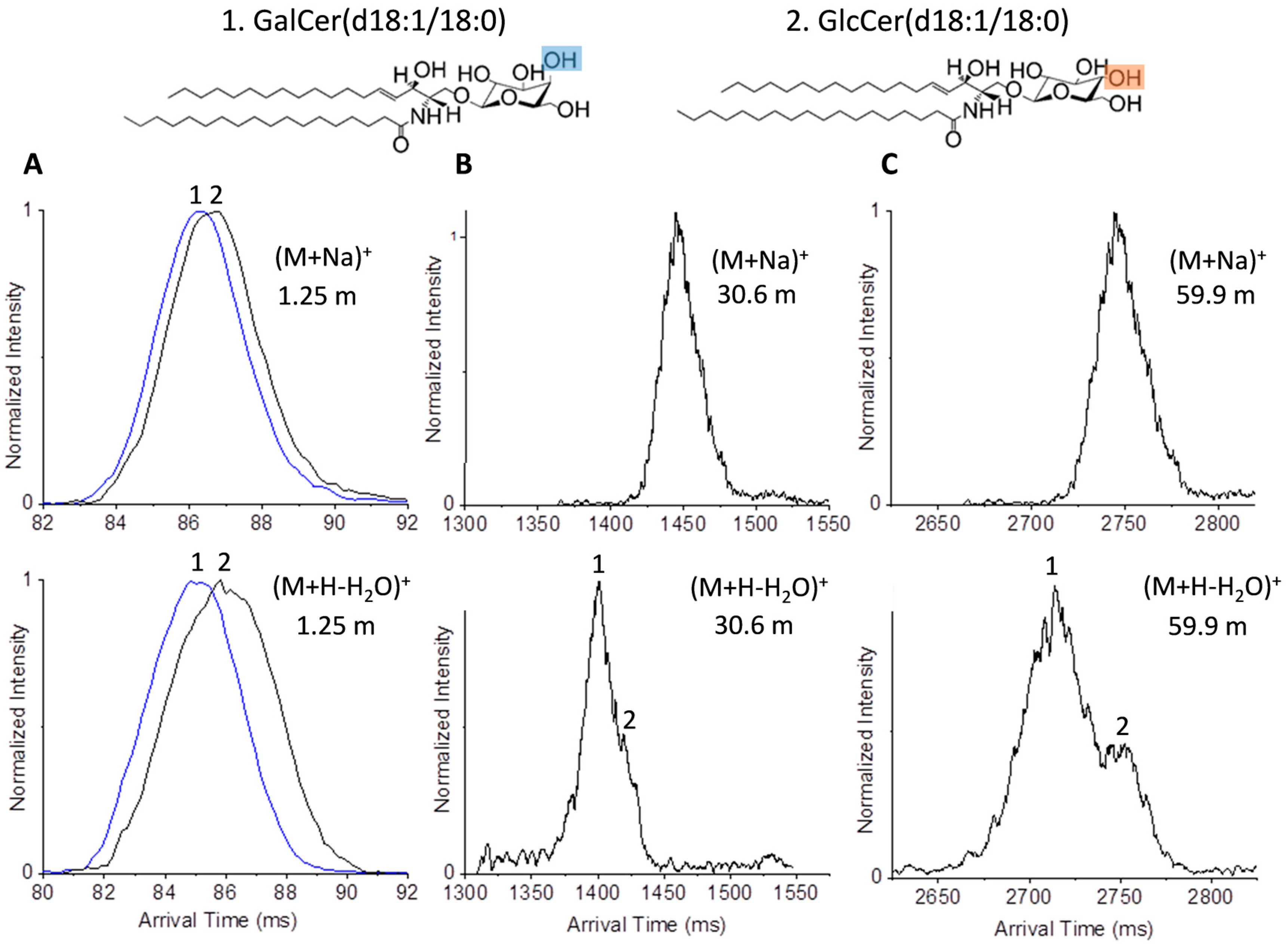
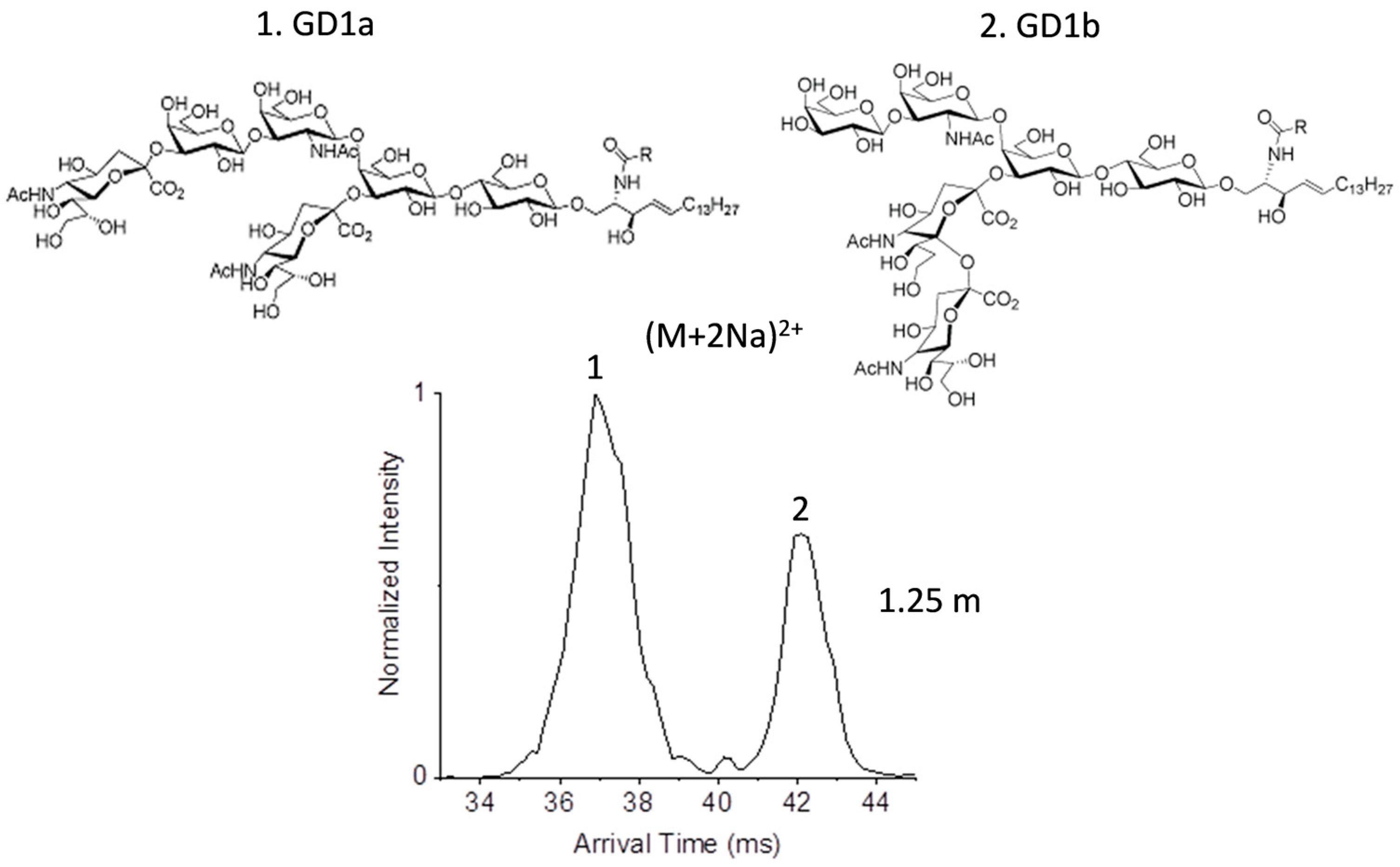
2.2. Glycolipid Isomers
3. Materials and Methods
3.1. Chemical Standards
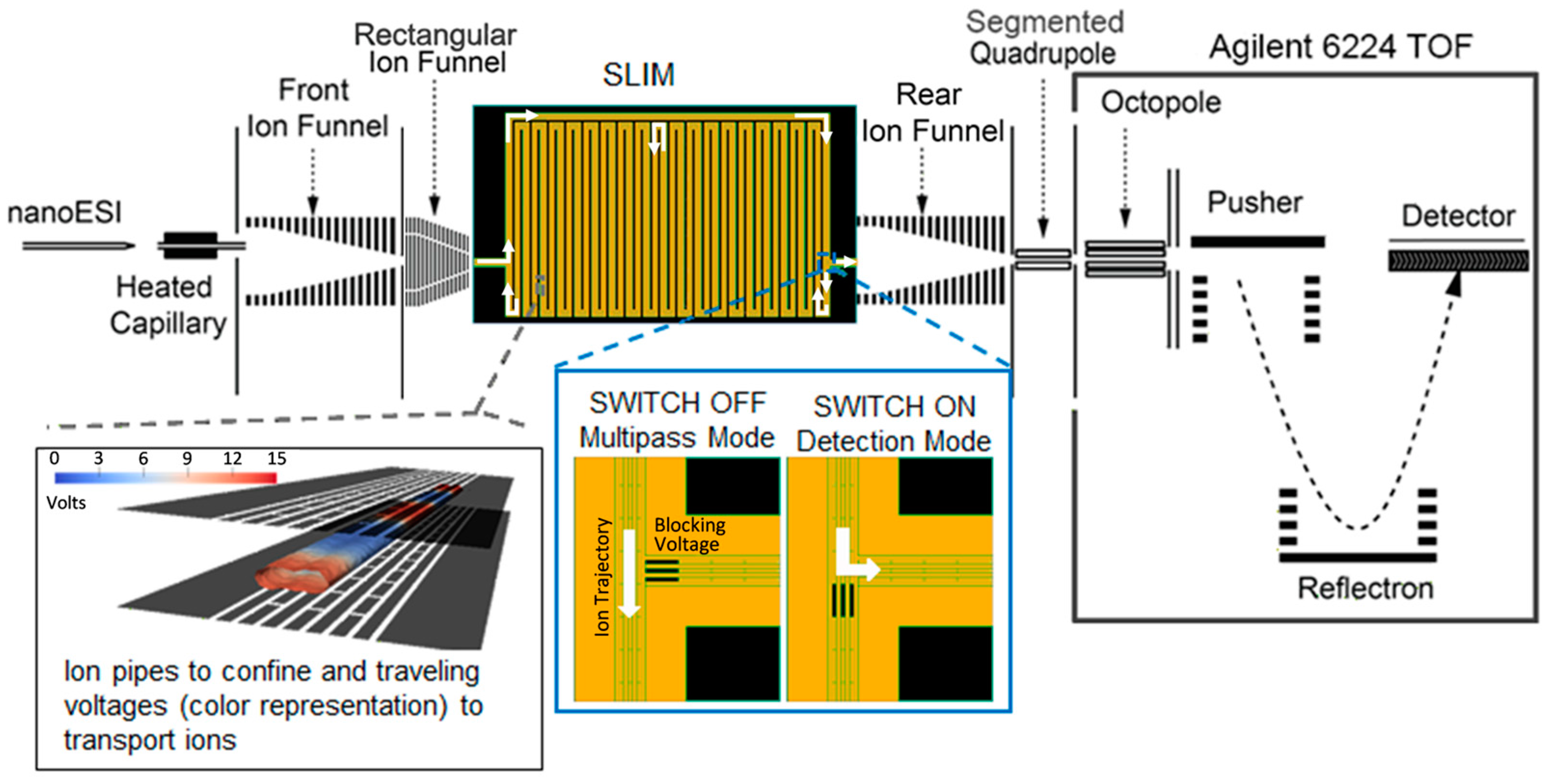
3.2. Instrumental Setup
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hullin-Matsuda, F.; Luquain-Costaz, C.; Bouvier, J.; Delton-Vandenbroucke, I. Bis(monoacylglycero)phosphate, a peculiar phospholipid to control the fate of cholesterol: Implications in pathology. Prostaglandins Leukot. Essent. Fat. Acids 2009, 81, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E.; Zhang, X.; Weitz, K.K.; Monroe, M.E.; Ibrahim, Y.M.; Moore, R.J.; Cha, J.; Sun, X.; Lovelace, E.S.; Wagoner, J.; et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst 2016, 141, 1649–1659. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.X.; Chong, L.; Tian, R.; Shi, R.Y.; Hu, T.Y.; Ouyang, Z.; Xia, Y. Identification and quantitation of lipid C=C location isomers: A shotgun lipidomics approach enabled by photochemical reaction. Proc. Natl. Acad. Sci. USA 2016, 113, 2573–2578. [Google Scholar] [CrossRef] [PubMed]
- Merrill, A.H., Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Daniotti, J.L.; Vilcaes, A.A.; Torres Demichelis, V.; Ruggiero, F.M.; Rodriguez-Walker, M. Glycosylation of glycolipids in cancer: Basis for development of novel therapeutic approaches. Front. Oncol. 2013, 3, 306. [Google Scholar] [CrossRef] [PubMed]
- Ledeen, R.W.; Wu, G. Nuclear lipids: Key signaling effectors in the nervous system and other tissues. J. Lipid Res. 2004, 45, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Daniotti, J.L.; Iglesias-Bartolome, R. Metabolic pathways and intracellular trafficking of gangliosides. IUBMB Life 2011, 63, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Maccioni, H.J. Glycosylation of glycolipids in the golgi complex. J. Neurochem. 2007, 103, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Tettamanti, G. Ganglioside/glycosphingolipid turnover: New concepts. Glycoconj. J. 2004, 20, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Mocchetti, I. Exogenous gangliosides, neuronal plasticity and repair, and the neurotrophins. Cell. Mol. Life Sci. 2005, 62, 2283–2294. [Google Scholar] [CrossRef] [PubMed]
- Nagle, J.F.; Tristram-Nagle, S. Structure of lipid bilayers. Biochim. Biophy. Acta 2000, 1469, 159–195. [Google Scholar] [CrossRef]
- Freudenberg, M.A.; Fomsgaard, A.; Mitov, I.; Galanos, C. Elisa for antibodies to lipid-A, lipopolysaccharides and other hydrophobic antigens. Infection 1989, 17, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Sokol, E.; Almeida, R.; Hannibal-Bach, H.K.; Kotowska, D.; Vogt, J.; Baumgart, J.; Kristiansen, K.; Nitsch, R.; Knudsen, J.; Ejsing, C.S. Profiling of lipid species by normal-phase liquid chromatography, nanoelectrospray ionization, and ion trap-orbitrap mass spectrometry. Anal. Biochem. 2013, 443, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Fauland, A.; Kofeler, H.; Trotzmuller, M.; Knopf, A.; Hartler, J.; Eberl, A.; Chitraju, C.; Lankmayr, E.; Spener, F. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J. Lipid Res. 2011, 52, 2314–2322. [Google Scholar] [CrossRef] [PubMed]
- Olsson, P.; Holmback, J.; Herslof, B. Separation of lipid classes by hplc on a cyanopropyl column. Lipids 2012, 47, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ioffe, V.; Kalendarev, T.; Rubinstein, I.; Zupkovitz, G. Reverse phase hplc for polar lipids. Simple and selective hplc procedures for analysis of phospholipid-based derivatives of valproic acid and various non-steroidal anti-inflammatory drugs. J. Pharm. Biomed. Anal. 2002, 30, 391–403. [Google Scholar] [CrossRef]
- Olsson, P.; Holmback, J.; Herslof, B. A single step reversed-phase high performance liquid chromatography separation of polar and non-polar lipids. J. Chromatogr. A 2014, 1369, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Morris, L.J. Separations of lipids by silver ion chromatography. J. Lipid Res. 1966, 7, 717–732. [Google Scholar] [PubMed]
- Dobson, G.; Christie, W.W.; Nikolova-Damyanova, B. Silver ion chromatography of lipids and fatty acids. J. Chromatogr. B Biomed. Appl. 1995, 671, 197–222. [Google Scholar] [CrossRef]
- Hughes, H.; Smith, C.V.; Horning, E.C.; Mitchell, J.R. High-performance liquid-chromatography and gas-chromatography mass-spectrometry determination of specific lipid-peroxidation products invivo. Anal. Biochem. 1983, 130, 431–436. [Google Scholar] [CrossRef]
- Fisk, H.L.; West, A.L.; Childs, C.E.; Burdge, G.C.; Calder, P.C. The use of gas chromatography to analyze compositional changes of fatty acids in rat liver tissue during pregnancy. J. Vis. Exp. 2014, 85, e51445. [Google Scholar] [CrossRef] [PubMed]
- Sandra, K.; Sandra, P. Lipidomics from an analytical perspective. Curr. Opin. Chem. Biol. 2013, 17, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.; Julian, R.R. Characterization of glycosphingolipid epimers by radical-directed dissociation mass spectrometry. Analyst 2016, 141, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Dilthey, B.G.; Gross, R.W. Identification and quantitation of fatty acid double bond positional isomers: A shotgun lipidomics approach using charge-switch derivatization. Anal. Chem. 2013, 85, 9742–9750. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.C.; Mitchell, T.W.; Harman, D.G.; Deeley, J.M.; Nealon, J.R.; Blanksby, S.J. Ozone-induced dissociation: Elucidation of double bond position within mass-selected lipid ions. Anal. Chem. 2008, 80, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.F.; Gross, M.L. Complete structural elucidation of triacylglycerols by tandem sector mass spectrometry. Anal. Chem. 1998, 70, 4417–4426. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, J.P.; Brodbelt, J.S. Structural characterization of gangliosides and glycolipids via ultraviolet photodissociation mass spectrometry. Anal. Chem. 2013, 85, 10399–10407. [Google Scholar] [CrossRef] [PubMed]
- Shvartsburg, A.A.; Isaac, G.; Leveque, N.; Smith, R.D.; Metz, T.O. Separation and classification of lipids using differential ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2011, 22, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; Goodwin, C.R.; Lareau, N.M.; Leaptrot, K.L.; Morris, C.B.; Kurulugama, R.T.; Mordehai, A.; Klein, C.; Barry, W.; Darland, E.; et al. Conformational ordering of biomolecules in the gas phase: Nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal. Chem. 2014, 86, 2107–2116. [Google Scholar] [CrossRef] [PubMed]
- Prost, S.A.; Crowell, K.L.; Baker, E.S.; Ibrahim, Y.M.; Clowers, B.H.; Monroe, M.E.; Gordon, A.; AndersonRichard, D.; SmithSamuel, H.P. Detecting and removing data artifacts in Hadamard transform ion mobility-mass spectrometry measurements. J. Am. Soc. Mass Spectrom. 2014, 25, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007. [Google Scholar] [CrossRef] [PubMed]
- Damen, C.W.; Isaac, G.; Langridge, J.; Hankemeier, T.; Vreeken, R.J. Enhanced lipid isomer separation in human plasma using reversed-phase UPLC with ion-mobility/high-resolution ms detection. J. Lipid Res. 2014, 55, 1772–1783. [Google Scholar] [CrossRef] [PubMed]
- Maccarone, A.T.; Duldig, J.; Mitchell, T.W.; Blanksby, S.J.; Duchoslav, E.; Campbell, J.L. Characterization of acyl chain position in unsaturated phosphatidylcholines using differential mobility-mass spectrometry. J. Lipid Res. 2014, 55, 1668–1677. [Google Scholar] [CrossRef] [PubMed]
- Groessl, M.; Graf, S.; Knochenmuss, R. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst 2015, 140, 6904–6911. [Google Scholar] [CrossRef] [PubMed]
- Hamid, A.M.; Ibrahim, Y.M.; Garimella, S.V.B.; Webb, I.K.; Deng, L.L.; Chen, T.C.; Anderson, G.A.; Prost, S.A.; Norheim, R.V.; Tolmachev, A.V.; et al. Characterization of traveling wave ion mobility separations in structures for loss less ion manipulations. Anal. Chem. 2015, 87, 11301–11308. [Google Scholar] [CrossRef] [PubMed]
- Webb, I.K.; Garimella, S.V.B.; Tolmachev, A.V.; Chen, T.C.; Zhang, X.Y.; Norheim, R.V.; Prost, S.A.; LaMarche, B.; Anderson, G.A.; Ibrahim, Y.M.; et al. Experimental evaluation and optimization of structures for loss less ion manipulations for ion mobility spectrometry with time-of-flight mass spectrometry. Anal. Chem. 2014, 86, 9169–9176. [Google Scholar] [CrossRef] [PubMed]
- Shvartsburg, A.A.; Smith, R.D. Fundamentals of traveling wave ion mobility spectrometry. Anal. Chem. 2008, 80, 9689–9699. [Google Scholar] [CrossRef] [PubMed]
- Giles, K.; Williams, J.P.; Campuzano, I. Enhancements in travelling wave ion mobility resolution. Rapid Commun. Mass Spectrom. 2011, 25, 1559–1566. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Ibrahim, Y.M.; Baker, E.S.; Aly, N.A.; Hamid, A.M.; Zhang, X.; Zheng, X.; Garimella, S.V.B.; Webb, I.K.; Prost, S.A.; et al. Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. ChemistrySelect 2016, 1, 2396–2399. [Google Scholar] [CrossRef]
- Chen, T.C.; Ibrahim, Y.M.; Webb, I.K.; Garimella, S.V.B.; Zhang, X.; Hamid, A.M.; Deng, L.L.; Karnesky, W.E.; Prost, S.A.; Sandoval, J.A.; et al. Mobility-selected ion trapping and enrichment using structures for lossless ion manipulations. Anal. Chem. 2016, 88, 1728–1733. [Google Scholar] [CrossRef] [PubMed]
- Garimella, S.V.B.; Ibrahim, Y.M.; Webb, I.K.; Ipsen, A.B.; Chen, T.C.; Tolmachev, A.V.; Baker, E.S.; Anderson, G.A.; Smith, R.D. Ion manipulations in structures for lossless ion manipulations (SLIM): Computational evaluation of a 90 degrees turn and a switch. Analyst 2015, 14, 6845–6852. [Google Scholar] [CrossRef] [PubMed]
- Webb, I.K.; Garimella, S.V.B.; Tolmachev, A.V.; Chen, T.C.; Zhang, X.Y.; Cox, J.T.; Norheim, R.V.; Prost, S.A.; LaMarche, B.; Anderson, G.A.; et al. Mobility-resolved ion selection in uniform drift field ion mobility spectrometry/mass spectrometry: Dynamic switching in structures for lossless ion manipulations. Anal. Chem. 2014, 86, 9632–9637. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Melchiorre, M.; Sansone, A.; Torreggiani, A. Lipid geometrical isomerism: From chemistry to biology and diagnostics. Chem. Rev. 2014, 114, 255–284. [Google Scholar] [CrossRef] [PubMed]
- Maggio, B.; Fanani, M.L.; Rosetti, C.M.; Wilke, N. Biophysics of sphingolipids ii. Glycosphingolipids: An assortment of multiple structural information transducers at the membrane surface. Biochim. Biophys. Acta 2006, 1758, 1922–1944. [Google Scholar] [CrossRef] [PubMed]
- Rahmann, H.; Rosner, H.; Kortje, K.H.; Beitinger, H.; Seybold, V. Ca2+-ganglioside-interaction in neuronal differentiation and development. Prog. Brain Res. 1994, 101, 127–145. [Google Scholar] [PubMed]
- Hakomori, S.I. Bifunctional role of glycosphingolipids—Modulators for transmembrane signaling and mediators for cellular interactions. J. Biol. Chem. 1990, 265, 18713–18716. [Google Scholar] [PubMed]
- Rolfs, A.; Giese, A.K.; Grittner, U.; Mascher, D.; Elstein, D.; Zimran, A.; Bottcher, T.; Lukas, J.; Hubner, R.; Golnitz, U.; et al. Glucosylsphingosine is a highly sensitive and specific biomarker for primary diagnostic and follow-up monitoring in gaucher disease in a non-jewish, caucasian cohort of gaucher disease patients. PLoS ONE 2013, 8, e79732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Igisu, H.; Suzuki, K. Progressive accumulation of toxic metabolite in a genetic leukodystrophy. Science 1984, 224, 753–755. [Google Scholar] [CrossRef] [PubMed]
- Han, X.L.; Cheng, H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 2005, 46, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Kerwin, J.L.; Watts, J.D.; Aebersold, R. Ceramide profiling of complex lipid mixtures by electrospray ionization mass spectrometry. Anal. Biochem. 1997, 244, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Haynes, C.A.; Allegood, J.C.; Park, H.; Sullards, M.C. Sphingolipidomics: Methods for the comprehensive analysis of sphingolipids. J. Chromatogr. B 2009, 877, 2696–2708. [Google Scholar] [CrossRef] [PubMed]
- Kusunoki, S.; Shimizu, J.; Chiba, A.; Ugawa, Y.; Hitoshi, S.; Kanazawa, I. Experimental sensory neuropathy induced by sensitization with ganglioside GD1B. Ann. Neurol. 1996, 39, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Kusunoki, S.; Chiba, A.; Kon, K.; Ando, S.; Arisawa, K.; Tate, A.; Kanazawa, I. N-acetylgalactosaminyl GD1A is a target molecule for serum antibody in guillain-barre-syndrome. Ann. Neurol. 1994, 35, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Ii, T.; Ohashi, Y.; Nagai, Y. Structural elucidation of underivatized gangliosides by electrospray-ionization tandem mass-spectrometry (ESIMS/MS). Carbohydr. Res. 1995, 273, 27–40. [Google Scholar] [CrossRef]
- Sarbu, M.; Robu, A.C.; Ghiulai, R.M.; Vukelic, Z.; Clemmer, D.E.; Zamfir, A.D. Electrospray ionization ion mobility mass spectrometry of human brain gangliosides. Anal. Chem. 2016, 88, 5166–5178. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.N.; Colsch, B.; Egan, T.; Lewis, E.K.; Schultz, J.A.; Woods, A.S. Gangliosides’ analysis by maldi-ion mobility ms. Analyst 2011, 136, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Ibrahim, Y.M.; Hamid, A.M.; Garimella, S.V.; Webb, I.K.; Zheng, X.; Prost, S.A.; Sandoval, J.A.; Norheim, R.V.; Anderson, G.A.; et al. Ultra-high resolution ion mobility separations utilizing traveling waves in a 13 m serpentine path length structures for lossless ion manipulations module. Anal. Chem. 2016, 88, 8957–8964. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.; Belov, M.E.; Tolmachev, A.V.; Prior, D.C.; Smith, R.D. Ion funnel trap interface for orthogonal time-of-flight mass spectrometry. Anal. Chem. 2007, 79, 7845–7852. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, Y.M.; Belov, M.E.; Liyu, A.V.; Smith, R.D. Automated gain control ion funnel trap for orthogonal time-offlight mass spectrometry. Anal. Chem. 2008, 80, 5367–5376. [Google Scholar] [CrossRef] [PubMed]
- Clowers, B.H.; Ibrahim, Y.M.; Prior, D.C.; Danielson, W.F., III; Belov, M.E.; Smith, R.D. Enhanced ion utilization efficiency using an electrodynamic ion funnel trap as an injection mechanism for ion mobility spectrometry. Anal. Chem. 2008, 80, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Webb, I.K.; Deng, L.; Hamid, A.M.; Anderson, G.A.; Norheim, R.V.; Prost, S.A.; Garimella, S.V.B.; Baker, E.S.; Ibrahim, Y.M.; Smith, R.D. Analysis of structural changes with high resolution structures for lossless ion manipulations (SLIM) ion mobility-mass spectrometry. In Proceedings of the 64th American Society for Mass Spectrometry Conference on Mass Spectrometry and Allied Topics, Indianapolis, Indiana, 4–8 June 2017, unpublished.
- Deng, L.; Ibrahim, Y.M.; Garimella, S.V.; Webb, I.K.; Hamid, A.M.; Norheim, R.V.; Prost, S.A.; Sandoval, J.A.; Baker, E.S.; Smith, R.D. Greatly increasing trapped ion populations for mobility separations using traveling waves in structures for lossless ion manipulations. Anal. Chem. 2016, 88, 10143–10150. [Google Scholar] [CrossRef] [PubMed]
- Garimella, S.V.; Ibrahim, Y.M.; Tang, K.; Webb, I.K.; Baker, E.S.; Tolmachev, A.V.; Chen, T.C.; Anderson, G.A.; Smith, R.D. Spatial ion peak compression and its utility in ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2016, 27, 1128–1135. [Google Scholar] [CrossRef] [PubMed]
- Garimella, S.V.B.; Hamid, A.M.; Deng, L.; Ibrahim, Y.M.; Webb, I.K.; Baker, E.S.; Prost, S.A.; Norheim, R.V.; Anderson, G.A.; Smith, R.D. The squeezing of ion populations and peaks in traveling wave ion mobility separations and structures for lossless ion manipulations using compression ratio ion mobility programming. Anal. Chem. 2016, 88, 11877–11885. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wojcik, R.; Webb, I.K.; Deng, L.; Garimella, S.V.B.; Prost, S.A.; Ibrahim, Y.M.; Baker, E.S.; Smith, R.D. Lipid and Glycolipid Isomer Analyses Using Ultra-High Resolution Ion Mobility Spectrometry Separations. Int. J. Mol. Sci. 2017, 18, 183. https://doi.org/10.3390/ijms18010183
Wojcik R, Webb IK, Deng L, Garimella SVB, Prost SA, Ibrahim YM, Baker ES, Smith RD. Lipid and Glycolipid Isomer Analyses Using Ultra-High Resolution Ion Mobility Spectrometry Separations. International Journal of Molecular Sciences. 2017; 18(1):183. https://doi.org/10.3390/ijms18010183
Chicago/Turabian StyleWojcik, Roza, Ian K. Webb, Liulin Deng, Sandilya V. B. Garimella, Spencer A. Prost, Yehia M. Ibrahim, Erin S. Baker, and Richard D. Smith. 2017. "Lipid and Glycolipid Isomer Analyses Using Ultra-High Resolution Ion Mobility Spectrometry Separations" International Journal of Molecular Sciences 18, no. 1: 183. https://doi.org/10.3390/ijms18010183




