Roles of GATA6 during Gonadal Development in Japanese Flounder: Gonadogenesis, Regulation of Gender-Related Genes, Estrogen Formation and Gonadal Function Maintenance
Abstract
:1. Introduction
2. Results
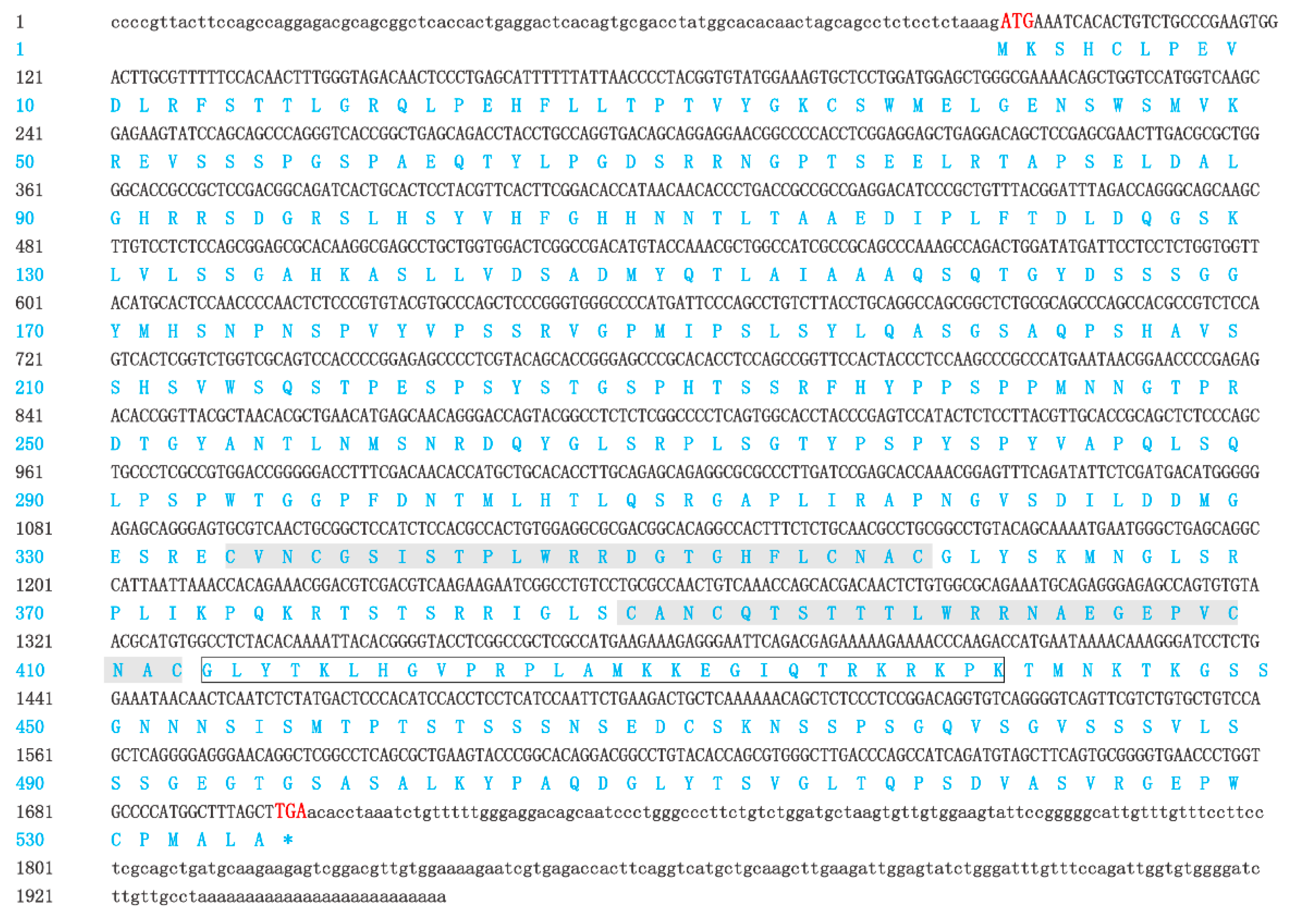
2.1. Isolation and Characterization of GATA6 cDNA
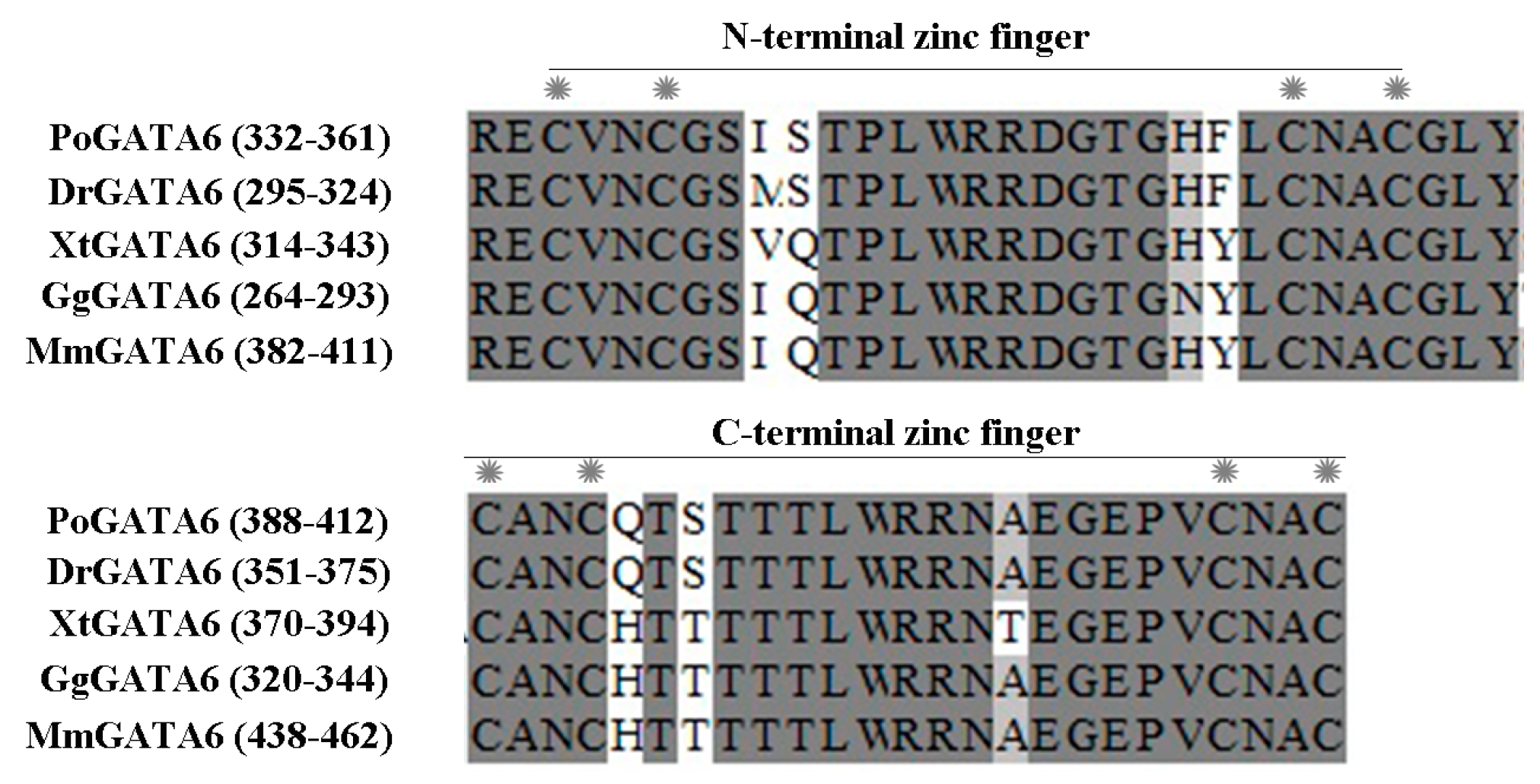
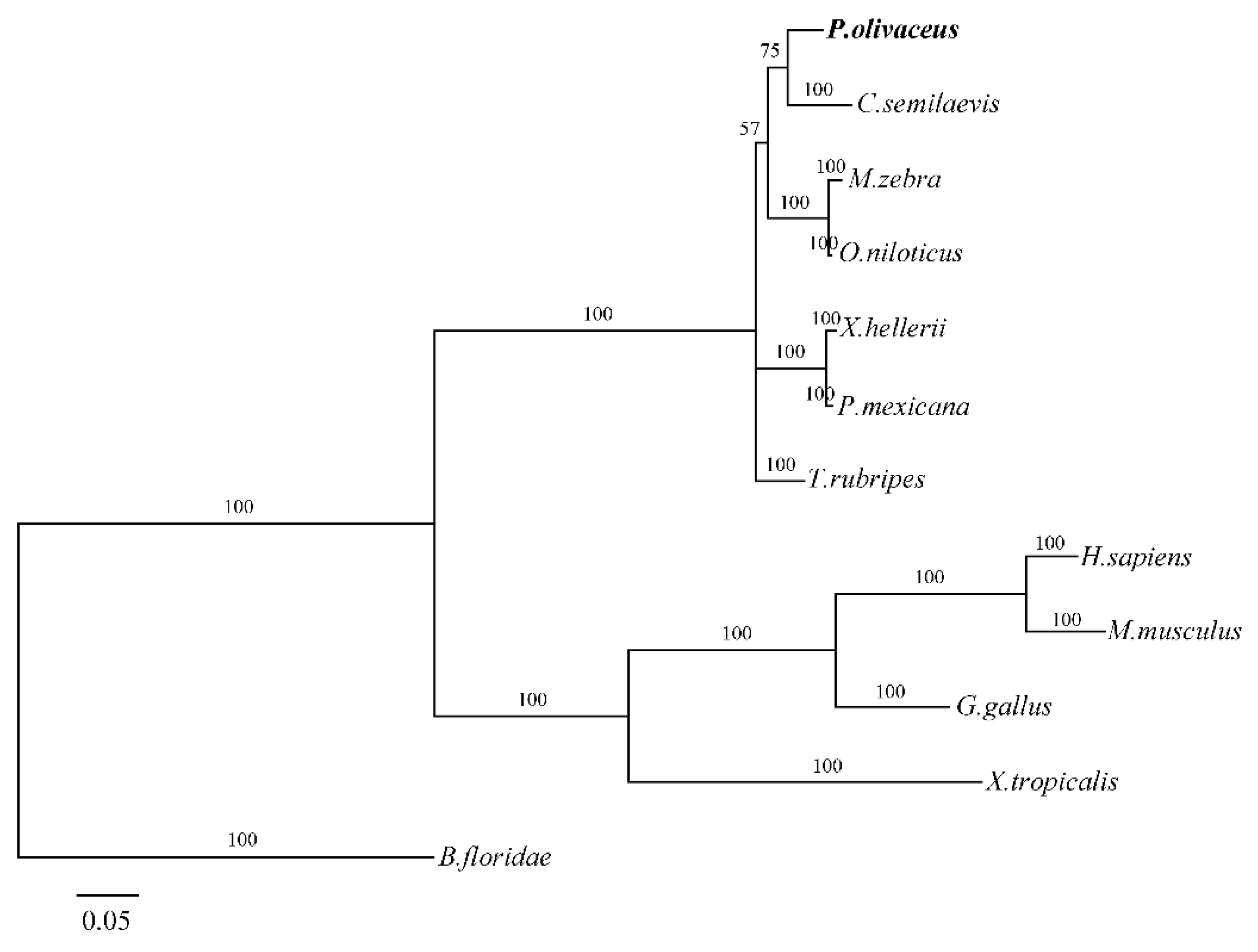
2.2. Homology and Phylogenetic Analysis
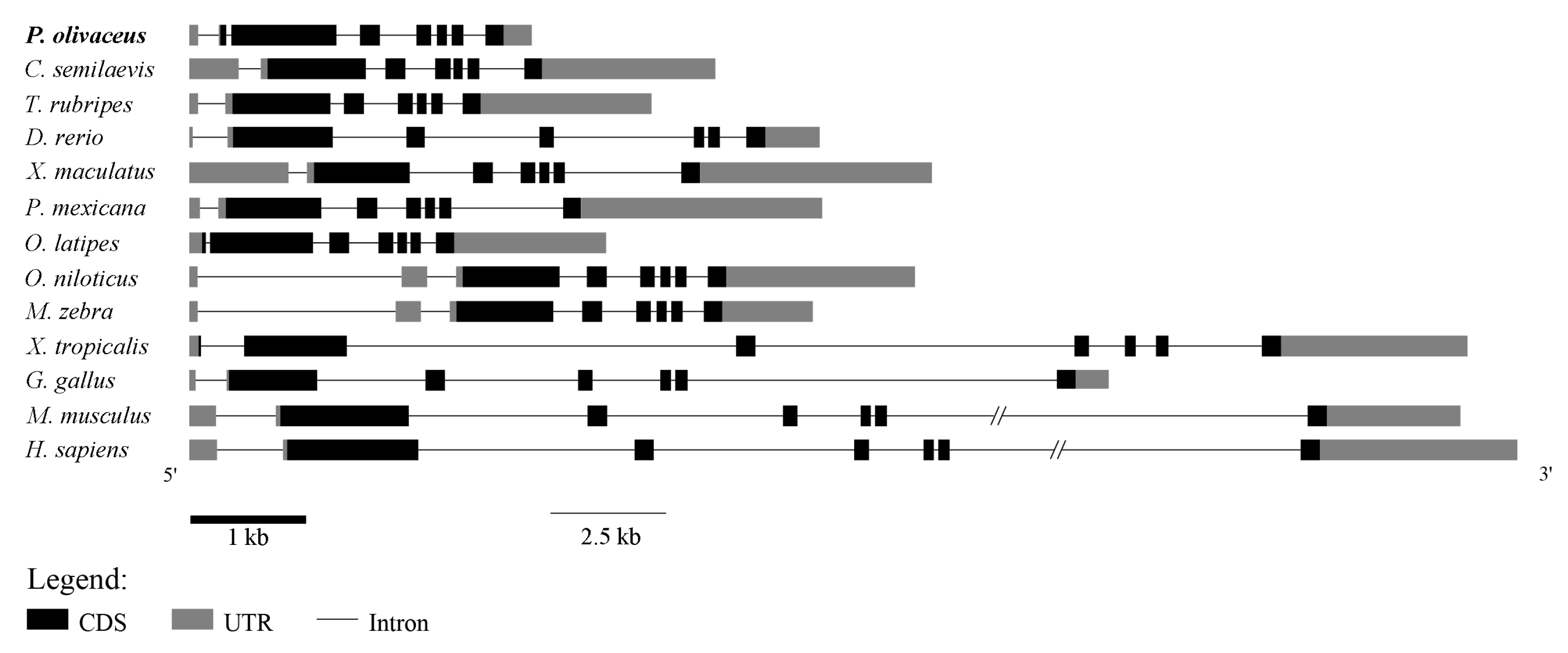
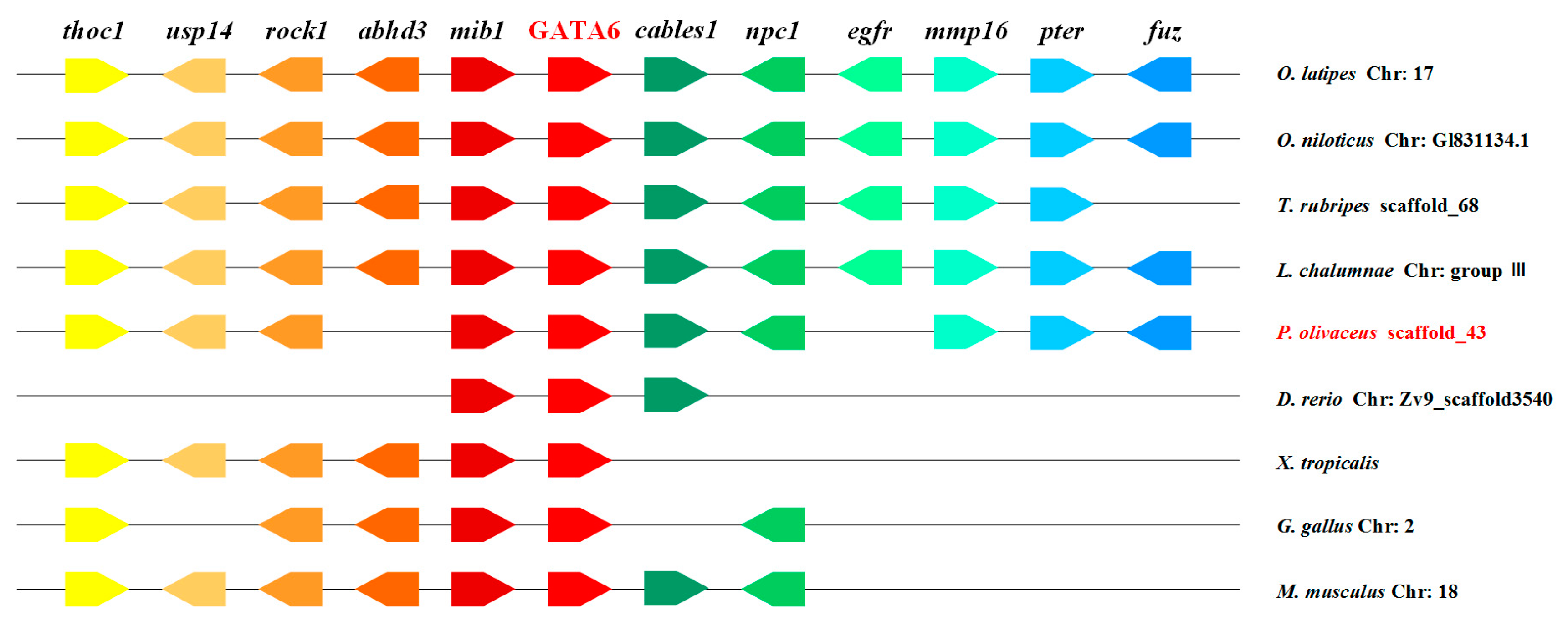
2.3. Genomic Synteny Analysis of GATA6
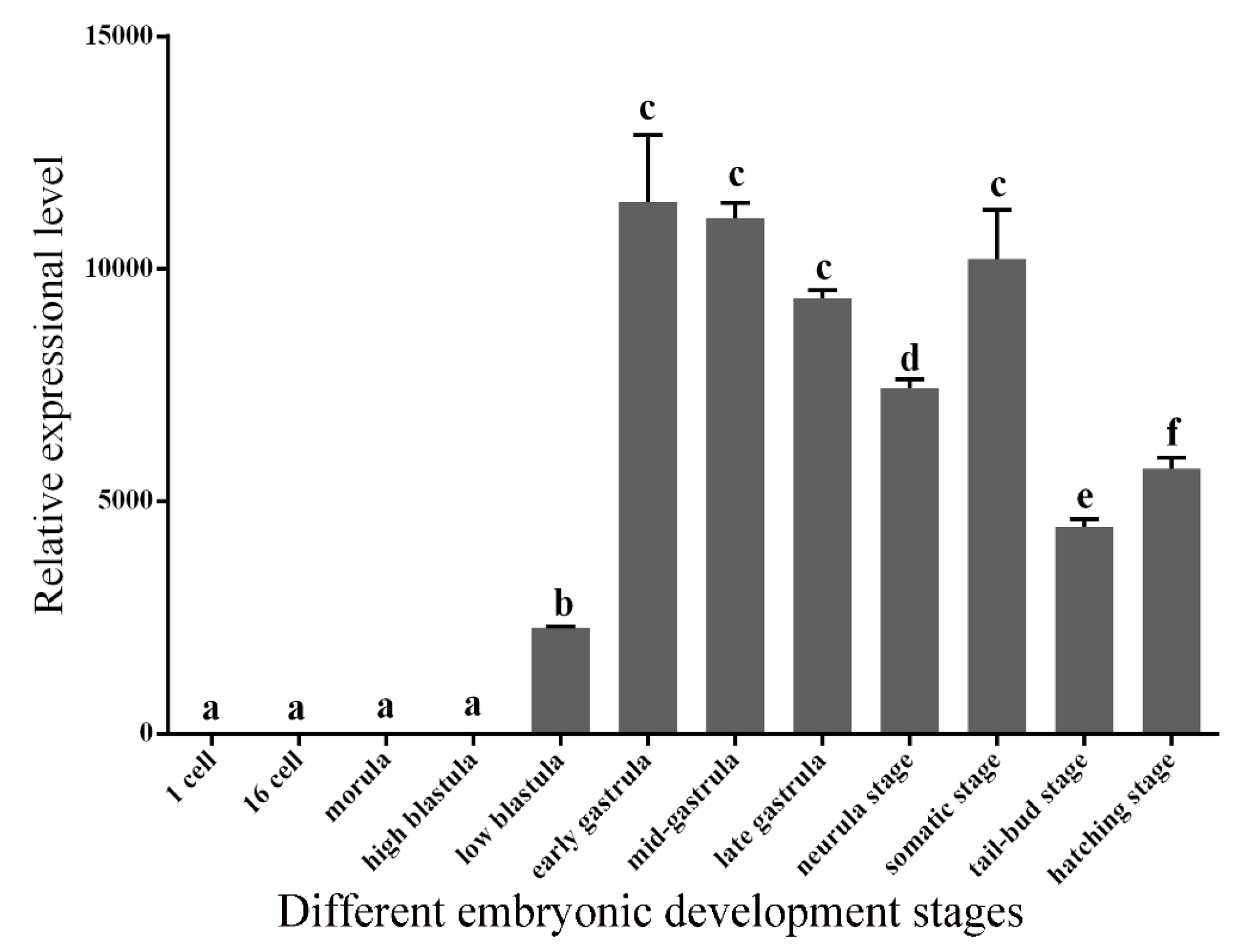
2.4. Expression Patterns of GATA6 at Different Embryonic Developmental Stages and in Different Tissues
2.5. In Situ Hybridization (ISH) of P. olivaceus GATA6 in Gonadal Sections
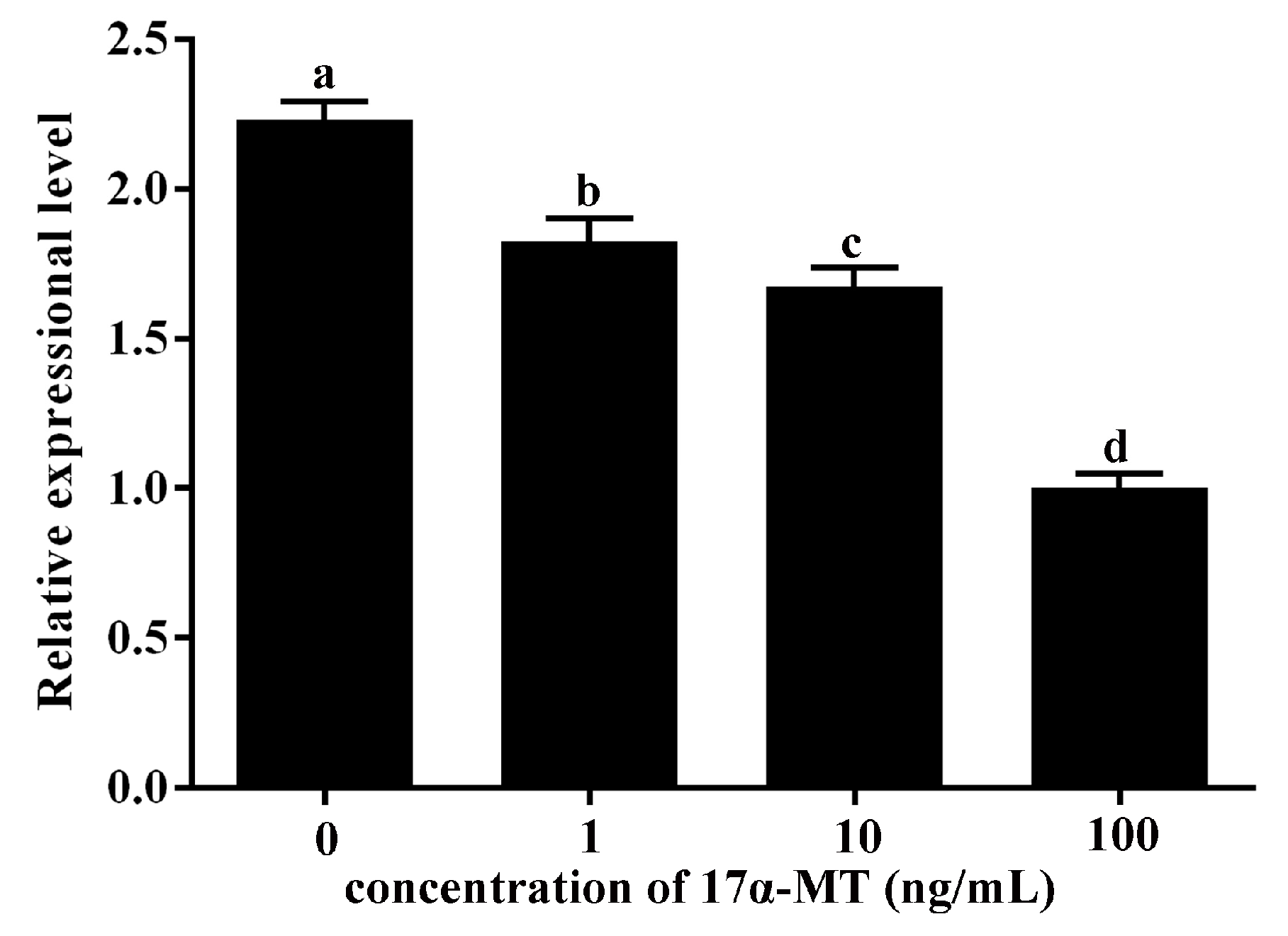
2.6. Expression Levels of GATA6 and CYP19A1 in Cultured P. olivaceus Testis Cells Stimulated with 17α-MT
3. Discussion
3.1. P. olivaceus GATA6 Forms the L-Type Protein
3.2. P. olivaceus GATA6 Is Highly Conserved in Vertebrates
3.3. P. olivaceus GATA6 Regulates Organs Originating from Endoderm and Mesoderm
3.4. P. olivaceus GATA6 Transcripts Are More Abundant in Testis than Ovary
3.5. P. olivaceus GATA6 Is Expressed in Oogonia, Oocytes and Sertoli Cells
3.6. P. olivaceus GATA6 Is Possibly Involved in the Synthesis of 17β Estradiol by Regulating CYP19A1
4. Materials and Methods
4.1. Ethics Statement
4.2. Specimen and Tissue Collection
4.3. RNA Extraction and cDNA Synthesis
4.4. Molecular Cloning of GATA6
4.5. Semi-Quantitative RT-PCR and qRT-PCR
4.6. ISH
4.7. Hormonal Stimulation and Primary Cultures of P. olivaceus Testis Cells
4.8. Bioinformatics Analysis and Phylogenetic Tree Construction
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lowry, J.A.; Atchley, W.R. Molecular evolution of the GATA family of transcription factors: Conservation within the DNA-binding domain. J. Mol. Evol. 2000, 50, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef]
- Lentjes, M.H.; Niessen, H.E.; Akiyama, Y.; de Bruine, A.P.; Melotte, V.; van Engeland, M. The emerging role of GATA transcription factors in development and disease. Expert Rev. Mol. Med. 2016, 18, e3. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.J.; Orkin, S.H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 1995, 23, 99–107. [Google Scholar] [PubMed]
- Huggon, I.C.; Davies, A.; Gove, C.; Moscoso, G.; Moniz, C.; Foss, Y.; Farzaneh, F.; Towner, P. Molecular cloning of human GATA6 DNA binding protein: High levels of expression in heart and gut. Biochim. Biophys. Acta 1997, 1353, 98–102. [Google Scholar] [CrossRef]
- Charron, F.; Nemer, M. GATA transcription factors and cardiac development. Semin. Cell Dev. Biol. 1999, 10, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.X.; Yang, J.Z.; Ai, Q.Y.; Miao, N.Z.; Zhao, Y.F.; Wang, Y.M. [The GATA family in reproduction]. Zhonghua Nan Ke Xue 2009, 15, 932–936. [Google Scholar] [PubMed]
- Laverriere, A.C.; MacNeill, C.; Mueller, C.; Poelmann, R.E.; Burch, J.B.; Evans, T. GATA4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994, 269, 23177–23184. [Google Scholar] [PubMed]
- Shu, J.; Zhang, K.; Zhang, M.; Yao, A.; Shao, S.; Du, F.; Yang, C.; Chen, W.; Wu, C.; Yang, W.; et al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res. 2015, 25, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Heikinheimo, M.; Ermolaeva, M.; Bielinska, M.; Rahman, N.A.; Narita, N.; Huhtaniemi, I.T.; Tapanainen, J.S.; Wilson, D.B. Expression and hormonal regulation of transcription factors GATA4 and GATA6 in the mouse ovary. Endocrinology 1997, 138, 3505–3514. [Google Scholar] [PubMed]
- Padua, M.B.; Fox, S.C.; Jiang, T.; Morse, D.A.; Tevosian, S.G. Simultaneous gene deletion of GATA4 and GATA6 leads to early disruption of follicular development and germ cell loss in the murine ovary. Biol. Reprod. 2014, 91, 24. [Google Scholar] [CrossRef] [PubMed]
- Convissar, S.M.; Bennett, J.; Baumgarten, S.C.; Lydon, J.P.; DeMayo, F.J.; Stocco, C. GATA4 and GATA6 Knockdown during Luteinization Inhibits Progesterone Production and Gonadotropin Responsiveness in the Corpus Luteum of Female Mice. Biol. Reprod. 2015, 93, 133. [Google Scholar] [CrossRef] [PubMed]
- Ketola, I.; Toppari, J.; Vaskivuo, T.; Herva, R.; Tapanainen, J.S.; Heikinheimo, M. Transcription factor GATA6, cell proliferation, apoptosis, and apoptosis-related proteins Bcl-2 and Bax in human fetal testis. J. Clin. Endocrinol. Metab. 2003, 88, 1858–1865. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.; Wu, Y.G.; Gossen, J.; Zhou, P.; Stocco, C. Loss of GATA6 and GATA4 in granulosa cells blocks folliculogenesis, ovulation, and follicle stimulating hormone receptor expression leading to female infertility. Endocrinology 2012, 153, 2474–2485. [Google Scholar] [CrossRef] [PubMed]
- Padua, M.B.; Jiang, T.; Morse, D.A.; Fox, S.C.; Hatch, H.M.; Tevosian, S.G. Combined loss of the GATA4 and GATA6 transcription factors in male mice disrupts testicular development and confers adrenal-like function in the testes. Endocrinology 2015, 156, 1873–1886. [Google Scholar] [CrossRef] [PubMed]
- Kiiveri, S.; Liu, J.; Westerholm-Ormio, M.; Narita, N.; Wilson, D.B.; Voutilainen, R.; Heikinheimo, M. Transcription factors GATA4 and GATA6 during mouse and human adrenocortical development. Endocr. Res. 2002, 28, 647–650. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, P.; Saner, K.; Mayhew, B.; Rainey, W.E. GATA6 is expressed in the human adrenal and regulates transcription of genes required for adrenal androgen biosynthesis. Endocrinology 2003, 144, 4285–4288. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, W.; Sun, Y.; Wang, Z.; Zhang, Q.; Wang, X. Molecular characterization and expression profiles of GATA6 in tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2016, 198, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D. The zinc finger-containing transcription factors GATA4, 5, and 6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 2000, 275, 38949–38952. [Google Scholar] [CrossRef] [PubMed]
- Pandian, T.J.; Sheela, S.G. Hormonal induction of sex reversal in fish. Aquaculture 1995, 138, 1–22. [Google Scholar] [CrossRef]
- Kitano, T.; Takamune, K.; Nagahama, Y.; Abe, S.I. Aromatase inhibitor and 17α-methyltestosterone cause sex-reversal from genetical females to phenotypic males and suppression of P450 aromatase gene expression in Japanese flounder (Paralichthys olivaceus). Mol. Reprod. Dev. 2000, 56, 1–5. [Google Scholar] [CrossRef]
- Bagheri-Fam, S.; Sinclair, A.H.; Koopman, P.; Harley, V.R. Conserved regulatory modules in the Sox9 testis-specific enhancer predict roles for SOX, TCF/LEF, Forkhead, DMRT, and GATA proteins in vertebrate sex determination. Int. J. Biochem. Cell Biol. 2010, 42, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Manuylov, N.L.; Fujiwara, Y.; Adameyko, I.I.; Poulat, F.; Tevosian, S.G. The regulation of Sox9 gene expression by the GATA4/FOG2 transcriptional complex in dominant XX sex reversal mouse models. Dev. Biol. 2007, 307, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Fujikura, J.; Yamato, E.; Yonemura, S.; Hosoda, K.; Masui, S.; Nakao, K.; Miyazaki, J.J.; Niwa, H. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002, 16, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Burch, J.B. Regulation of GATA gene expression during vertebrate development. Semin. Cell Dev. Biol. 2005, 16, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Takada, K.; Obayashi, K.; Ohashi, K.; Ohashi-Kobayashi, A.; Nakanishi-Matsui, M.; Maeda, M. Amino-terminal extension of 146 residues of L-type GATA6 is required for transcriptional activation but not for self-association. Biochem. Biophys. Res. Commun. 2014, 452, 962–966. [Google Scholar] [CrossRef] [PubMed]
- Maeda, M.; Ohashi, K.; Ohashi-Kobayashi, A. Further extension of mammalian GATA6. Dev. Growth Differ. 2005, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Obayashi, K.; Kobayashi, A.; Maeda, M. A unique role of an amino terminal 16-residue region of long-type GATA6. J. Biochem. 2004, 135, 639–650. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Xia, L.; Dong, X.; Hu, S.; Zhang, Y.; Ding, F.; Liu, H.; Li, L.; Wang, J. Transcription factors GATA4 and GATA6: Molecular characterization, expression patterns and possible functions during goose (Anser cygnoides) follicle development. J. Reprod. Dev. 2014, 60, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, W.; Du, X.; Jiang, J.; Wang, C.; Wang, X.; Zhang, Q.; He, Y. Molecular characterization and functional analysis of the GATA4 in tongue sole (Cynoglossus semilaevis). Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2016, 193, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gillio-Meina, C.; Hui, Y.Y.; LaVoie, H.A. GATA4 and GATA6 transcription factors: Expression, immunohistochemical localization, and possible function in the porcine ovary. Biol. Reprod. 2003, 68, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Brewer, A.; Gove, C.; Davies, A.; McNulty, C.; Barrow, D.; Koutsourakis, M.; Farzaneh, F.; Pizzey, J.; Bomford, A.; Patient, R. The human and mouse GATA6 genes utilize two promoters and two initiation codons. J. Biol. Chem. 1999, 274, 38004–38016. [Google Scholar] [CrossRef] [PubMed]
- Gillis, W.Q.; St, J.J.; Bowerman, B.; Schneider, S.Q. Whole genome duplications and expansion of the vertebrate GATA transcription factor gene family. BMC Evol. Biol. 2009, 9, 207. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.A. Generation of embryos directly from embryonic stem cells by tetraploid embryo complementation reveals a role for GATA factors in organogenesis. Biochem. Soc. Trans. 2005, 33, 1534–1536. [Google Scholar] [CrossRef] [PubMed]
- Capo-Chichi, C.D.; Smedberg, J.L.; Rula, M.; Nicolas, E.; Yeung, A.T.; Adamo, R.F.; Frolov, A.; Godwin, A.K.; Xu, X.X. Alteration of Differentiation Potentials by Modulating GATA Transcription Factors in Murine Embryonic Stem Cells. Stem Cells Int. 2010, 2010, 602068. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Sakurai, T.; Godkin, J.D.; Imakawa, K. Expression and potential role of GATA factors in trophoblast development. J. Reprod. Dev. 2013, 59, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ketola, I.; Rahman, N.; Toppari, J.; Bielinska, M.; Porter-Tinge, S.B.; Tapanainen, J.S.; Huhtaniemi, I.T.; Wilson, D.B.; Heikinheimo, M. Expression and regulation of transcription factors GATA4 and GATA6 in developing mouse testis. Endocrinology 1999, 140, 1470–1480. [Google Scholar] [PubMed]
- LaVoie, H.A. The GATA-keepers of ovarian development and folliculogenesis. Biol. Reprod. 2014, 91, 38. [Google Scholar] [CrossRef] [PubMed]
- Paul-Prasanth, B.; Bhandari, R.K.; Kobayashi, T.; Horiguchi, R.; Kobayashi, Y.; Nakamoto, M.; Shibata, Y.; Sakai, F.; Nakamura, M.; Nagahama, Y. Estrogen oversees the maintenance of the female genetic program in terminally differentiated gonochorists. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Jiang, X.; Xie, Q.; Yuan, J.; Huang, B.; Tao, W.; Zhou, L.; Nagahama, Y.; Wang, D. Transdifferentiation of Differentiated Ovary into Functional Testis by Long-Term Treatment of Aromatase Inhibitor in Nile Tilapia. Endocrinology 2014, 155, 1476–1488. [Google Scholar] [CrossRef] [PubMed]
- Takatsu, K.; Miyaoku, K.; Roy, S.R.; Murono, Y.; Sago, T.; Itagaki, H.; Nakamura, M.; Tokumoto, T. Induction of Female-to-Male Sex Change in Adult Zebrafish by Aromatase Inhibitor Treatment. Sci. Rep. 2013, 3. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Xu, Y.; Wang, Y.; Wei, S.; Feng, D.; Huang, Q.; Zhang, S.; Liu, Z. Developmental expression and immune role of the class B scavenger receptor CD36 in zebrafish. Dev. Comp. Immunol. 2016, 60, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, J.; Jiang, J.; Fan, L.; Wang, W.; Liu, J.; Zhang, Q.; Wang, X. Identification and characterization of a nanog homolog in Japanese flounder (Paralichthys olivaceus). Gene 2013, 531, 411–421. [Google Scholar] [CrossRef] [PubMed]



| Primers | Sequence (5′–3′) | Tm (°C) | Usage |
|---|---|---|---|
| gata6-Fw | GRTCAAGSGAGAAGTATCC | 59 | Degenerate PCR |
| gata6-Rv | CWGATRGCTGGGTCAAG | 59 | Degenerate PCR |
| gata6-5′Rv1 | GCTGGATACTTCTCGCTTGAC | 63 | 5′ RACE |
| gata6-5′Rv2 | CCGTGGTTGGTGGTCATTAT | 62 | 5′ RACE |
| gata6-3′Fw1 | CGCTCGCCATGAAGAAAGA | 62 | 3′ RACE |
| gata6-3′Fw2 | ACAAAGGGATCCTCTGGAAATAA | 62 | 3′ RACE |
| gata6-RT-Fw | CATCCACCTCCTCATCCAATTC | 62 | qRT-PCR/semi-RT-PCR |
| gata6-RT-Rv | CATCTGATGGCTGGGTCAAG | 62 | qRT-PCR/semi-RT-PCR |
| Cyp19a1-RT-Rv | CACCTTTCTGTTTGGGTTTG | 62 | qRT-PCR |
| Cyp19a1-RT-Rv | GTCTCCATACCTCTTGTTGTAG | 62 | qRT-PCR |
| gata6-ISH-Fw | CATCCACCTCCTCATCCAATTC | 62 | ISH probe |
| gata6-ISH-Rv | CCACACCAATCTGGAAACAAATC | 62 | ISH probe |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Liu, X.; Sun, Y.; Liu, J.; Liu, Y.; Wang, M.; Zhang, Q.; Wang, X. Roles of GATA6 during Gonadal Development in Japanese Flounder: Gonadogenesis, Regulation of Gender-Related Genes, Estrogen Formation and Gonadal Function Maintenance. Int. J. Mol. Sci. 2017, 18, 160. https://doi.org/10.3390/ijms18010160
Li Z, Liu X, Sun Y, Liu J, Liu Y, Wang M, Zhang Q, Wang X. Roles of GATA6 during Gonadal Development in Japanese Flounder: Gonadogenesis, Regulation of Gender-Related Genes, Estrogen Formation and Gonadal Function Maintenance. International Journal of Molecular Sciences. 2017; 18(1):160. https://doi.org/10.3390/ijms18010160
Chicago/Turabian StyleLi, Zan, Xiumei Liu, Yan Sun, Jinxiang Liu, Yuezhong Liu, Mengxun Wang, Quanqi Zhang, and Xubo Wang. 2017. "Roles of GATA6 during Gonadal Development in Japanese Flounder: Gonadogenesis, Regulation of Gender-Related Genes, Estrogen Formation and Gonadal Function Maintenance" International Journal of Molecular Sciences 18, no. 1: 160. https://doi.org/10.3390/ijms18010160






