Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain
Abstract
:1. Introduction
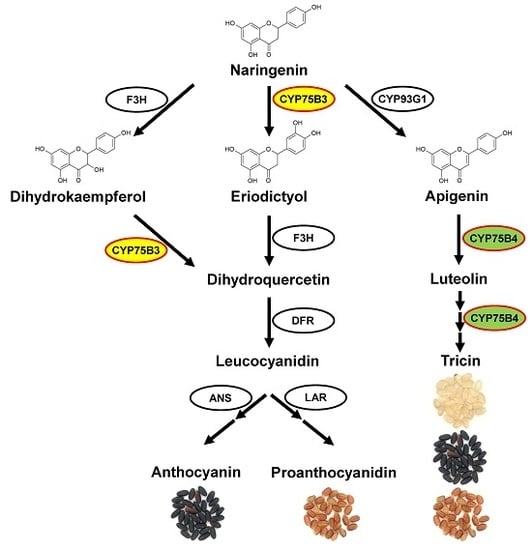
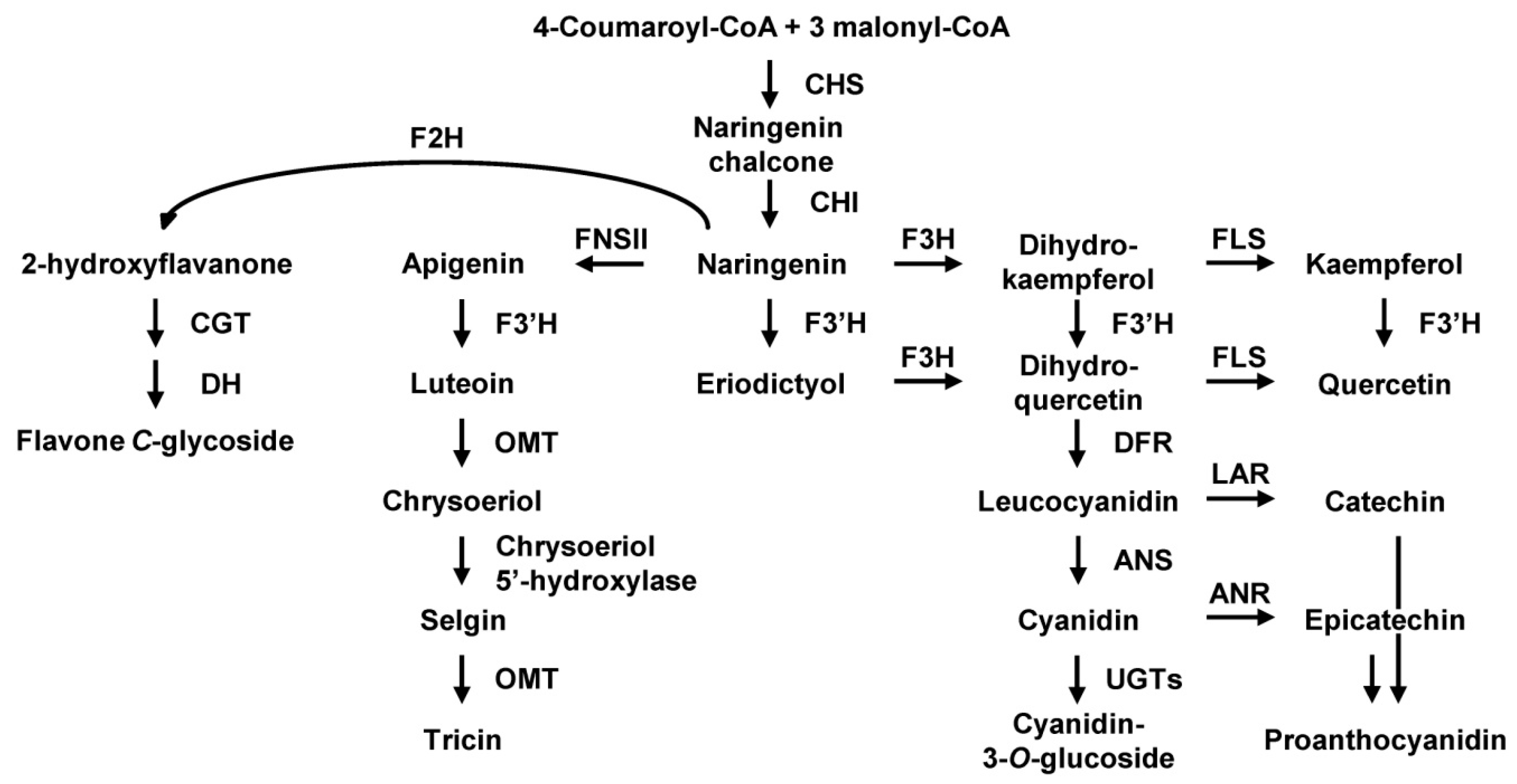
2. Results
2.1. Sequence Analysis of CYP75B3s and CYP75B4s
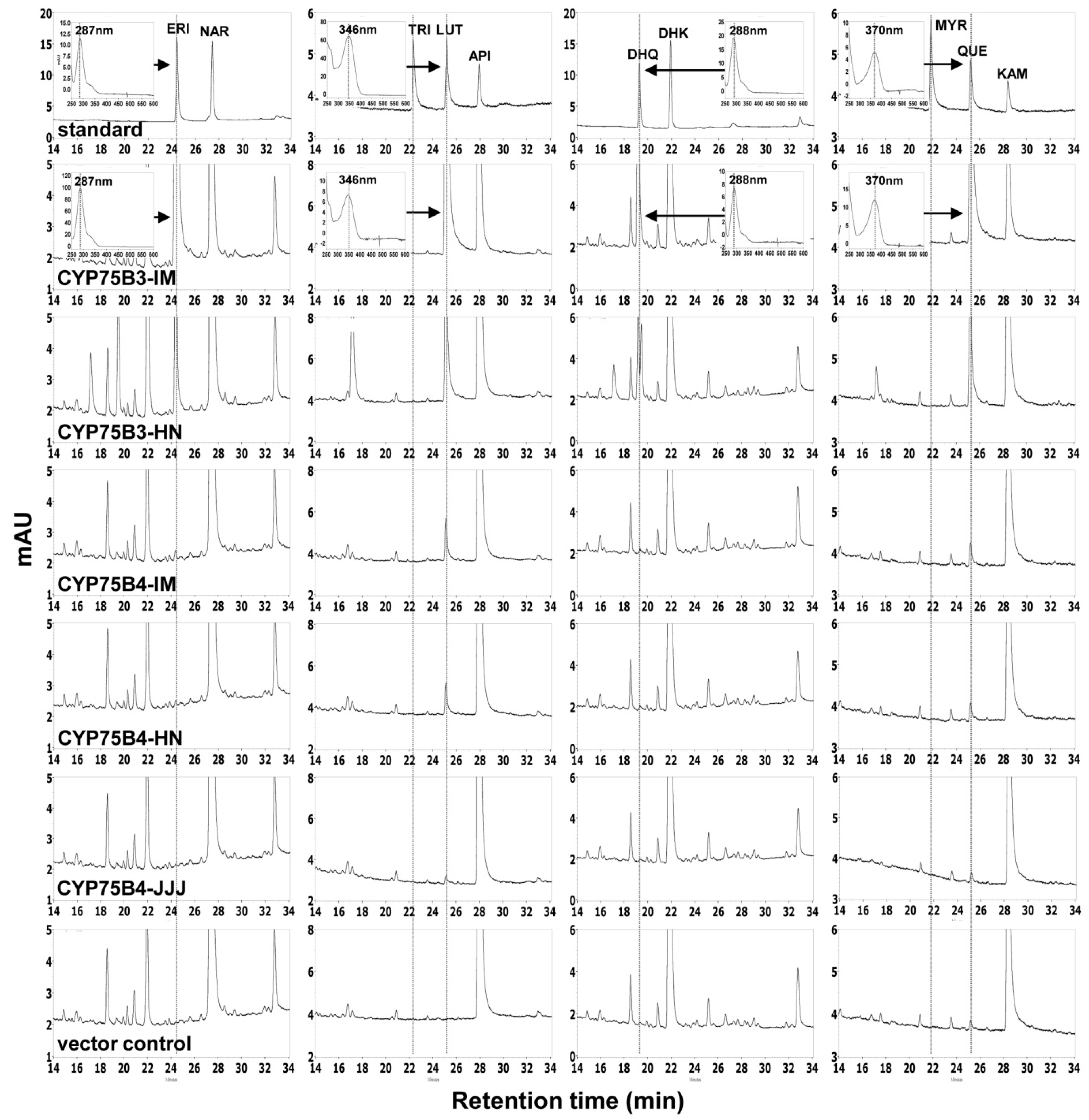
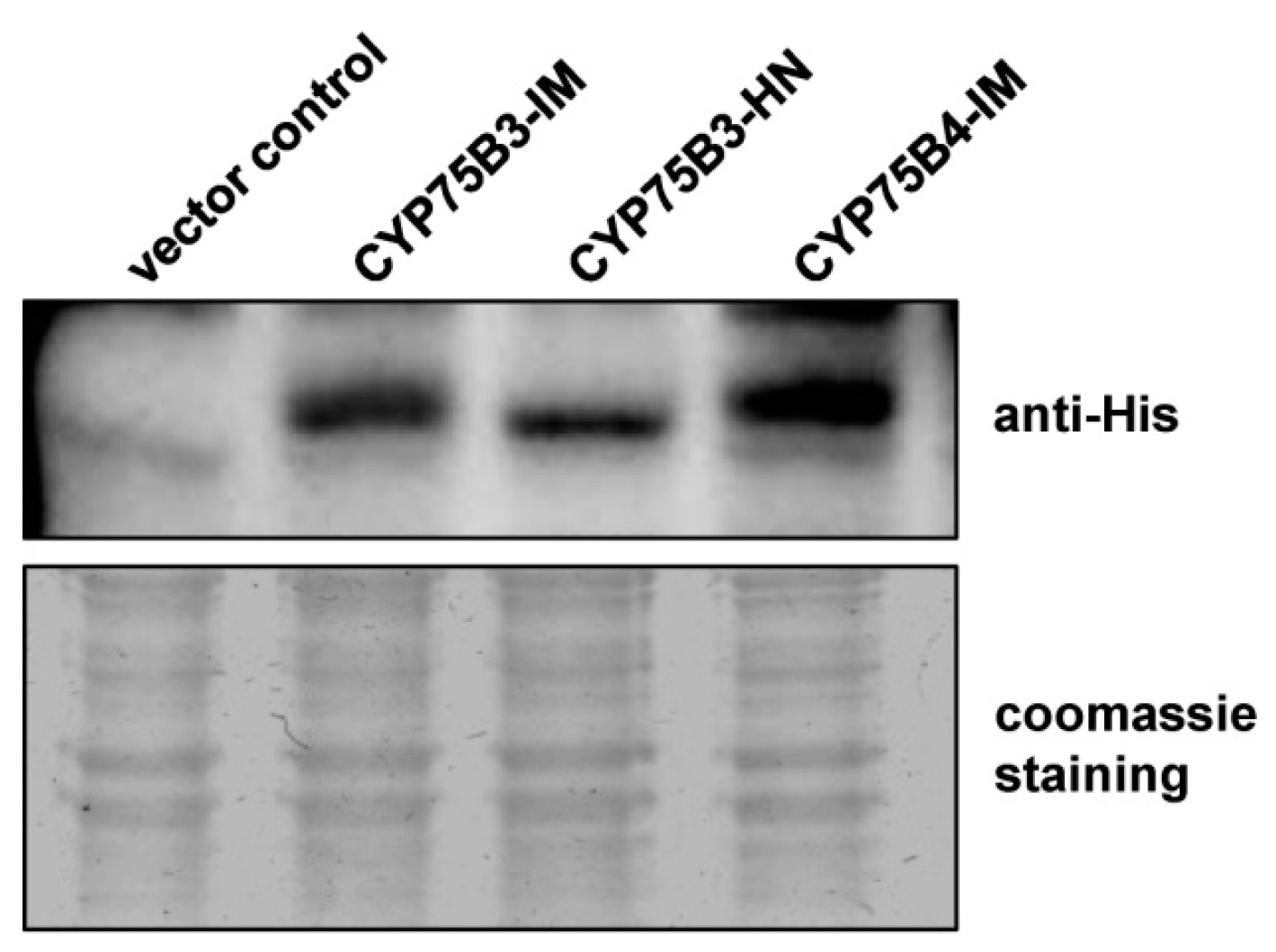
2.2. Yeast Expression of CYP75Bs and CYP75B4s and Enzyme Assays
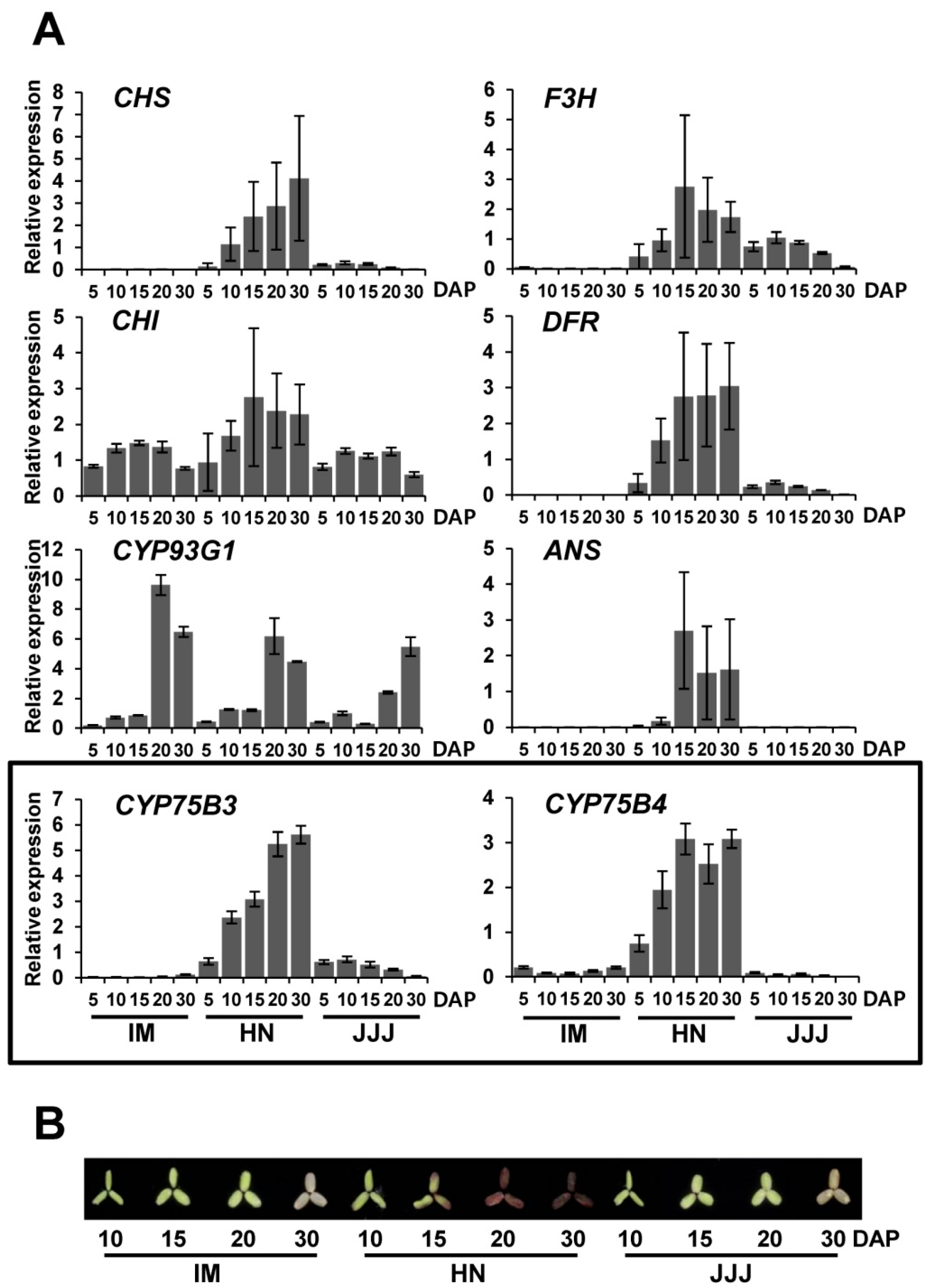
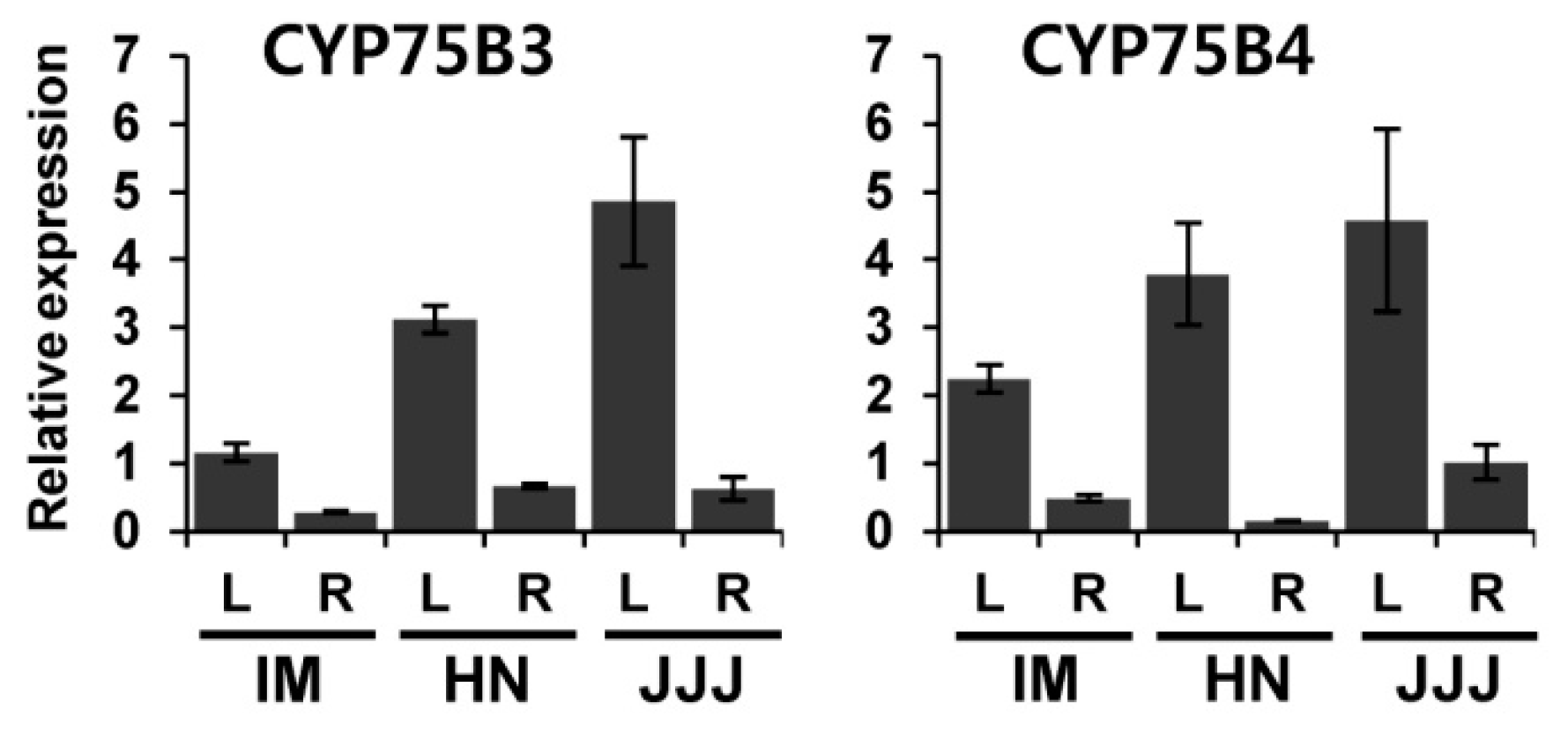
2.3. Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Isolation of CYP75B3 and CYP75B4 cDNAs from Non-Pigmented and Pigmented Rice
4.3. Yeast Expression of CYP75B3s and CYP75B4s and Substrate Feeding Assay
4.4. Microsomal Protein Preparation
4.5. Standard Enzyme Assay
4.6. HPLC Analysis
4.7. Immunoblot Analysis
4.8. Gene Expression Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hu, C.; Zawistowski, J.; Ling, W.; Kitts, D.D. Black rice (Oryza sativa L. indica) pigmented fraction suppresses both reactive oxygen species and nitric oxide in chemical and biological model systems. J. Agric. Food Chem. 2003, 51, 5271–5277. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.D.; Ling, W.H.; Ma, J.; Xia, M.; Hou, M.J.; Wang, Q.; Zhu, H.L.; Tang, Z.H. An anthocyanin-rich extract from black rice enhances atherosclerotic plaque stabilization in apolipoprotein E-deficientmice. J. Nutr. 2006, 136, 2220–2225. [Google Scholar] [PubMed]
- Cho, M.K.; Kim, M.H.; Kang, M.Y. Effects of rice embryo and embryo jelly with black rice bran pigment on lipid metabolism and antioxidant enzyme activity in high cholesterol-fed rats. J. Korean Soc. Appl. Biol. Chem. 2008, 51, 200–206. [Google Scholar]
- Finocchiaro, F.; Ferrari, B.; Gianinetti, A.; Dall′asta, C.; Galaverna, G.; Scazzina, F.; Pellegrini, N. Characterization of antioxidant compounds of red and white rice and changes in total antioxidant capacity during processing. Mol. Nutr. Food Res. 2007, 51, 1006–1019. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.N.; Kuo, W.H.; Chiang, C.L.; Chiou, H.L.; Hsieh, Y.S.; Chu, S.C. Black rice anthocyanins inhibit cancer cells invasion via repressions of MMPs and u-PA expression. Chem. Biol. Interact. 2006, 163, 218–229. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Mi, M.; Ling, W. Effects of anthocyanin-rich extract from black rice alone or combined with chemotherapeutic agents on proliferation of different cancer cells. Acta Acad. Med. Militaris Tertiae 2007, 29, 1943–1946. [Google Scholar]
- Hyun, J.W.; Yang, Y.M.; Sung, M.S.; Chung, H.S.; Paik, W.H.; Kang, S.S.; Park, J.G. The cytotoxic activity of sterol erivatives from Pulsatilla chinensis Regal. J. Korean Cancer Assoc. 1996, 28, 145–151. [Google Scholar]
- Kim, H.M.; Kang, C.S.; Lee, E.H.; Shin, T.Y. The evaluation of the antianaphylactic effect of Oryza sativa L. subsp. hsien Ting in rats. Pharmacol. Res. 1999, 40, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.P.; Kang, M.Y.; Koh, H.J.; Nam, S.H.; Friedman, M. Antiallergic activities of pigmented rice bran extracts in cell assays. J. Food Sci. 2007, 72, S719–S726. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Xu, H.; Zheng, Y.X.; Xu, X.; Wan, F.J.; Zhou, X.M. Effect of anthocyanin-rich extract from black rice on the survival of mouse brain neuron. Cell Biol. Int. 2008. [Google Scholar] [CrossRef]
- Abdel-Aal, E.-S.M.; Young, J.C.; Rabalski, I. Anthocyanin composition in black, blue, pink, purple, and red cereal grains. J. Agric. Food Chem. 2006, 54, 4696–4704. [Google Scholar] [CrossRef] [PubMed]
- Nakornriab, M.; Sriseadka, T.; Wongpornchai, S. Quantification of carotenoid and flavonoid components in brans of some Thai black rice cultivars using supercritical fluid extraction and high-performance liquid chromatography-mass spectrometry. J. Food Lipids 2008, 15, 488–503. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, S.Y.; Chu, S.M.; Lim, S.H.; Suh, S.C.; Lee, Y.T.; Cho, H.S.; Ha, S.H. Variation and correlation analysis of flavonoids and carotenoids in Korean pigmented rice (Oryza sativa L.) cultivars. J. Agric. Food Chem. 2010, 58, 12804–12809. [Google Scholar] [CrossRef] [PubMed]
- Pereira-Caro, G.; Watanabe, S.; Crozier, A.; Fujimura, T.; Yokota, T.; Ashihara, H. Phytochemical profile of a Japanese black-purple rice. Food Chem. 2013, 141, 2821–2827. [Google Scholar] [CrossRef] [PubMed]
- Bordiga, M.; Gomez-Alonso, S.; Locatelli, M.; Travaglia, F.; Coïsson, J.D.; Hermosin-Gutierrez, I.; Arlorio, M. Phenolics characterization and antioxidant activity of six different pigmented Oryza sativa L. cultivars grown in Piedmont (Italy). Food Res. Int. 2014, 65, 282–290. [Google Scholar] [CrossRef]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012, 35, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Goufo, P.; Trindade, H. Rice antioxidants: Phenolic acids, flavonoids, anthocyanins, proanthocyanidins, tocopherols, tocotrienols, γ-oryzanol, and phytic acid. Food Sci. Nutr. 2014, 2, 75–104. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Ozawa, K.; Ishimaru, T.; Murayama, T.; Takaiwa, F. Transgenic rice seed synthesizing diverse flavonoids at high levels: A new platform for flavonoid production with associated health benefits. Plant Biotechnol. J. 2013, 11, 734–746. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Liu, H.; Lo, C. Completion of tricin biosynthesis pathway in rice: Cytochrome P450 75B4 is a unique chrysoeriol 5′-hydroxylase. Plant Physiol. 2015, 168, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Renuka Devi, R.; Jayalekshmy, A.; Arumughan, C. Antioxidant efficacy of phytochemical extracts from defatted rice bran in the bulk oil system. Food Chem. 2007, 104, 658–664. [Google Scholar] [CrossRef]
- Li, M.; Pu, Y.; Yoo, C.G.; Ragauskas, A.J. The occurrence of tricin and its derivatives in plants. Green Chem. 2016, 18, 1439–1454. [Google Scholar] [CrossRef]
- Shih, C.H.; Chu, H.; Tang, L.K.; Sakamoto, W.; Maekawa, M.; Chu, I.K.; Wang, M.; Lo, C. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 2008, 228, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Yamaguchi, T.; Omoteno, M.; Takarada, T.; Fujita, K.; Murata, K.; Iyama, Y.; Kojima, Y.; Morikawa, M.; Ozaki, H.; et al. Genetic dissection of black grain rice by the development of a near isogenic line. Breed. Sci. 2014, 64, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, T.; Maeda, H.; Oguchi, T.; Yamaguchi, T.; Tanabe, N.; Ebana, K.; Yano, M.; Ebitani, T.; Izawa, T. The birth of a black rice gene and its local spread by introgression. Plant Cell 2015, 27, 2401–2414. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, T.; Maekawa, M.; Oki, T.; Suda, I.; Iida, S.; Shimada, H.; Takamure, I.; Kadowaki, K. The Rc and Rd genes are involved in proanthocyanidin synthesis in rice pericarp. Plant J. 2007, 49, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Seitz, C.; Ameres, S.; Forkmann, G. Identification of the molecular basis for the functional difference between flavonoid 3′-hydroxylase and flavonoid 3′,5′-hydroxylase. FEBS Lett. 2007, 581, 3429–3434. [Google Scholar] [CrossRef] [PubMed]
- Thill, J.; Miosic, S.; Gotame, T.P.; Mikulic-Petkovsek, M.; Gosch, C.; Veberic, R.; Preuss, A.; Schwab, W.; Stampar, F.; Stich, K.; et al. Differential expression of flavonoid 3′-hydroxylase during fruit development establishes the different B-ring hydroxylation patterns of flavonoids in Fragaria × ananassa and Fragaria vesca. Plant Physiol Biochem. 2013, 72, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.S.; Zhou, R.; Yu, Y.B.; Xiao, Y.; Li, D.H.; Xiao, B.; Yu, O.; Yang, Y.J. Cloning and characterization of a flavonoid 3′-hydroxylase gene from tea plant (Camellia sinensis). Int. J. Mol. Sci. 2016. [Google Scholar] [CrossRef] [PubMed]
- Schlangen, K.; Miosic, S.; Halbwirth, H. Allelic variants from Dahlia variabilis encode flavonoid 3′-hydroxylases with functional differences in chalcone 3-hydroxylase activity. Arch. Biochem. Biophys. 2010, 494, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, Y.J.; Kim, B.G.; Lim, Y.; Ahn, J.H. Flavanone 3β-hydroxylases from rice: Key enzymes for favonol and anthocyanin biosynthesis. Mol. Cells 2008, 25, 312–316. [Google Scholar] [PubMed]
- Winkel-Shirley, B. Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol. Plant. 1999, 107, 142–149. [Google Scholar] [CrossRef]
- Galland, M.; Boutet-Mercey, S.; Lounifi, I.; Godin, B.; Balzergue, S.; Grandjean, O.; Morin, H.; Perreau, F.; Debeaujon, I.; Rajjou, L. Compartmentation and dynamics of flavone metabolism in dry and germinated rice seeds. Plant Cell Physiol. 2014, 55, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Zhu, F.Y.; Chan, W.L.; Liu, H.; Lo, C. Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol. 2014, 165, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chu, H.; Chu, I.K.; Lo, C. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 2010, 154, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Pompon, D.; Louerat, B.; Bronine, A.; Urban, P. Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 1996, 272, 51–64. [Google Scholar] [PubMed]
- Gietz, D.; St Jean, A.; Woods, R.A.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1425. Available online: http://www.ncbi.nlm.nih.gov/pubmed/1561104 (accessed on 5 August 2016). [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lim, S.H.; Ha, S.H. Marker development for the identification of rice seed color. Plant Biotechnol. Rep. 2013, 7, 391–398. [Google Scholar] [CrossRef]

| OsF3′Hs | Substrates | ||||
|---|---|---|---|---|---|
| NAR | API | DHK | KAM | ||
| CYP75B3-IM (CYP75B3-JJJ) | Km (µM) | 0.286 ± 0.015 | 0.072 ± 0.011 | 2.494 ± 0.350 | 0.110 ± 0.008 |
| Vmax (μM∙min−1·mg−1) | 0.214 ± 0.015 | 0.131 ± 0.011 | 0.430 ± 0.05 | 0.240 ± 0.014 | |
| Vmax/Km (1∙min−1·mg−1) | 0.746 ± 0.067 | 1.809 ± 0.168 | 0.173 ± 0.018 | 2.178 ± 0.282 | |
| CYP75B3-HN | Km (μM) | 0.108 ± 0.009 | 0.085 ± 0.015 | 1.034 ± 0.107 | 0.098 ± 0.013 |
| Vmax (μM∙min−1·mg−1) | 0.193 ± 0.029 | 0.161 ± 0.008 | 0.281 ± 0.023 | 0.270 ± 0.021 | |
| Vmax/Km (1∙min−1·mg−1) | 1.796 ± 0.240 | 1.887 ± 0.156 | 0.271 ± 0.034 | 2.755 ± 0.323 | |
| CYP75B4-IM | Km (μM) | ND | 0.240 ± 0.042 | * | 2.948 ± 0.330 |
| Vmax (μM∙min−1·mg−1) | ND | 0.008 ± 0.001 | * | 0.007 ± 0.001 | |
| Vmax/Km (1∙min−1·mg−1) | ND | 0.034 ± 0.003 | * | 0.002 ± 0.001 | |
| Target | Locus ID | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|---|
| qPCR | |||
| CYP75B3 | Os10g0320100 | ACGGATTCATCAACGAAAGG | AGCAGCACGCTTAGAAGGTC |
| CYP75B4 | Os10g0317900 | TCTCCCATCCGCTTACAATA | ACCAATCTACCAACATACAACAA |
| CHS | Os11g0530600 | GGGCTCATCTCGAAGAACAT * | CCTCATCCTCTCCTTGTCCA * |
| CHI | Os03g0819600 | AATCGAGCTGCGAATTAACC * | CGCGATTTCTCCTTTCCTTT * |
| FNSII | Os04g0101400 | GATTGGCAGTGCATGGACA | TTGTATTACGGTGCGTTACAGG |
| F3H | Os04g0662600 | AGCACAGAAGCCCAAGTCTC * | CTTCGATTTTCGACGGAAGA * |
| DFR | Os01g0633500 | GCGAGAAGGAACCGATACTG * | TCCAAATCTCGCATTGTGAA * |
| ANS | Os01g0372500 | GCATCGAACGGAATGAGAAC * | TTCGCTTCCGTTGAACATTA * |
| UBQ5 | Os01g0328400 | GAAGTAAGGAAGGAGGAGGA * | AAGGTGTTCAGTTCCAAGG * |
| Cloning | |||
| CYP75B3 | CACCATGGACGTTGTGCCTCTCCCGC | TACTCCATAAGCCGATGGAAGCAGC | |
| CYP75B4 | CACCATGGAGGTCGCCGCCATGG | TGCAATATTGTAAGCGGATGGGAGAAG |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Choi, M.J.; Lee, J.Y.; Kim, J.K.; Ha, S.-H.; Lim, S.-H. Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain. Int. J. Mol. Sci. 2016, 17, 1549. https://doi.org/10.3390/ijms17091549
Park S, Choi MJ, Lee JY, Kim JK, Ha S-H, Lim S-H. Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain. International Journal of Molecular Sciences. 2016; 17(9):1549. https://doi.org/10.3390/ijms17091549
Chicago/Turabian StylePark, Sangkyu, Min Ji Choi, Jong Yeol Lee, Jae Kwang Kim, Sun-Hwa Ha, and Sun-Hyung Lim. 2016. "Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain" International Journal of Molecular Sciences 17, no. 9: 1549. https://doi.org/10.3390/ijms17091549
APA StylePark, S., Choi, M. J., Lee, J. Y., Kim, J. K., Ha, S.-H., & Lim, S.-H. (2016). Molecular and Biochemical Analysis of Two Rice Flavonoid 3’-Hydroxylase to Evaluate Their Roles in Flavonoid Biosynthesis in Rice Grain. International Journal of Molecular Sciences, 17(9), 1549. https://doi.org/10.3390/ijms17091549