Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism
Abstract
:1. Introduction
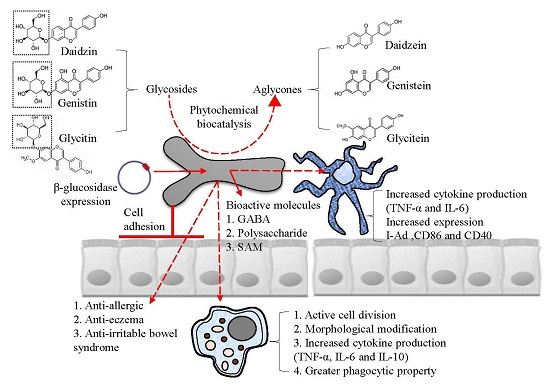
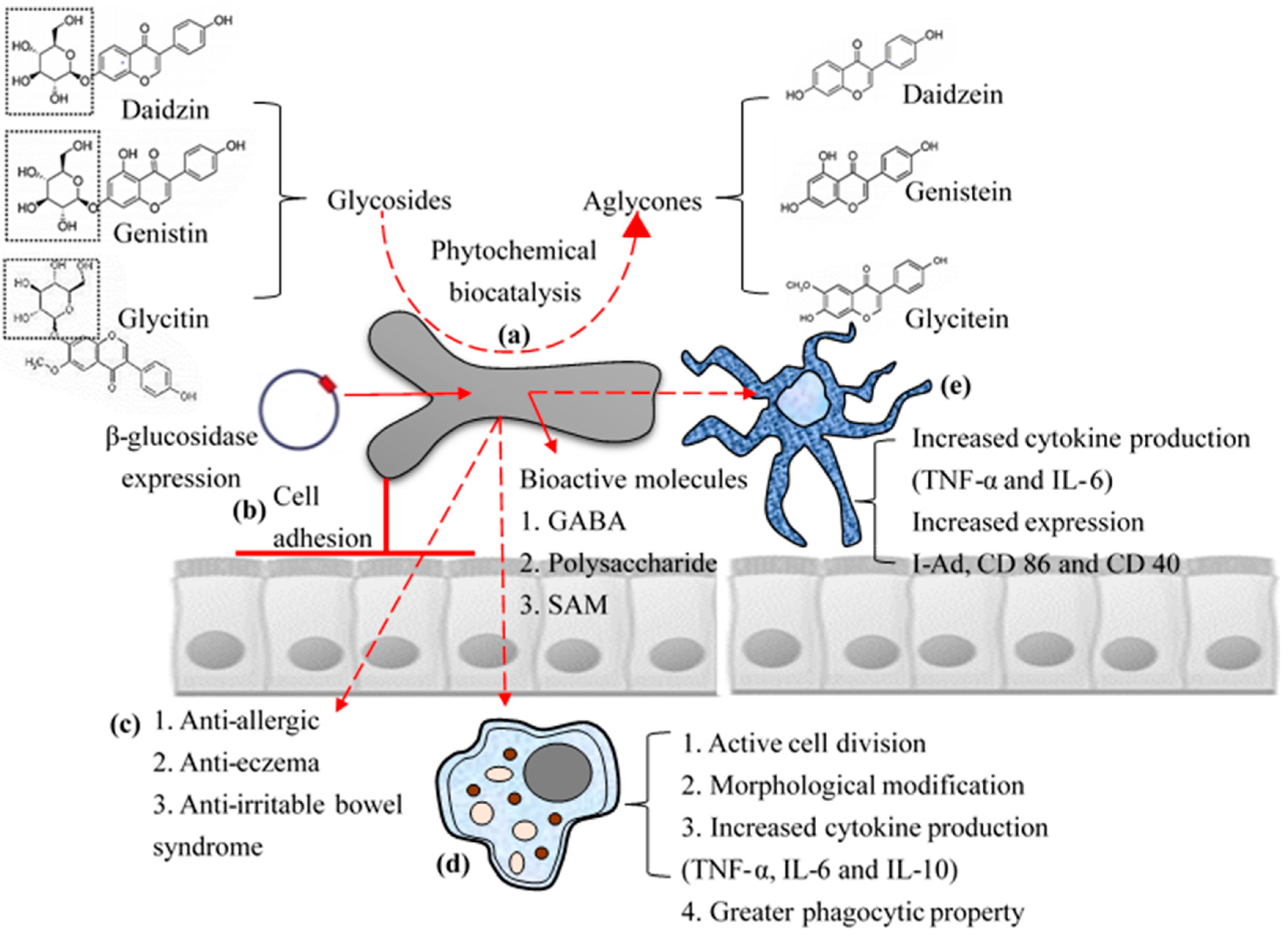
2. Cell Adhesive Property
3. Immune-Modulatory Effects of B. bifidum BGN4
4. Anticancer Effects of B. bifidum BGN4
5. Industrial Application: Biocatalysis
6. Industrial Application: Bioactive Molecules
7. Increase Biomass Productivity
8. From Comparative Genomics to Functionality of BGN4
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| BGN4 | Bifidobacterium bifidum BGN4 |
| CD | cluster of differentiation: |
| FDA | USA Food and Drug Administration |
| GABA | γ-Aminobutyric acid |
| IFN | Interferon |
| IL | Interleukin |
| SAM | S-Adenosyl-l-Methionine |
| SEM | scanning electron microscope |
| TNF | tumor necrosis factor |
References
- Siró, I.; Kápolna, E.; Kápolna, B.; Lugasi, A. Functional food. Product development, marketing and consumer acceptance: A review. Appetite 2008, 51, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Syngai, G.; Gopi, R.; Bharali, R.; Dey, S.; Lakshmanan, G.; Ahmed, G. Probiotics—The versatile vunctional food ingredients. J. Food Sci. Technol. 2015, 53, 921–933. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration, Labeling & Nutrition. Available online: http://www.fda.gov/Food/IngredientsPackagingLabeling/LabelingNutrition/ (accessed on 30 June 2016).
- De Prisco, A.; Mauriello, G. Probiotication of foods: A focus on microencapsulation tool. Trends Food Sci. Technol. 2016, 48, 27–39. [Google Scholar] [CrossRef]
- Yildiz, F. Development and Manufacture of Yogurt and Other Functional Dairy Products; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2010; pp. 1–36. [Google Scholar]
- Preedy, V. Handbook of Diet, Nutrition and The Skin; Wageningen Academic Publisher: Wageningen, The Netherland, 2012; pp. 327–328. [Google Scholar]
- Sarkar, S.; Sur, A.; Sarkar, K.; Majhi, R.; Basu, S.; Chatterjee, K.; Sikder, B. Probiotics: A way of value addition in functional food. Int. J. Food Sci. Nutr. Diet. 2016, 5, 290–293. [Google Scholar]
- Tajabadi, N.; Ebrahimpour, A.; Baradaran, A.; Rahim, R.; Mahyudin, N.; Manap, M.; Bakar, F.; Saari, N. Optimization Of γ-Aminobutyric acid production by Lactobacillus plantarum Taj-Apis362 from honeybees. Molecules 2015, 20, 6654–6669. [Google Scholar] [CrossRef] [PubMed]
- Ku, S. Finding and producing probiotic glycosylases for the biocatalysis of ginsenosides: A mini review. Molecules 2016, 21, 645. [Google Scholar] [CrossRef] [PubMed]
- Soto, A.; Martín, V.; Jiménez, E.; Mader, I.; Rodríguez, J.; Fernández, L. Lactobacilli and bifidobacteria in human bBreast milk: Influence of antibiotherapy and other host and clinical factors. J. Pediatr. Gastroenterol. Nutr. 2014, 59, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Arboleya, S.; Ruas-Madiedo, P.; Margolles, A.; Solís, G.; Salminen, S.; Clara, G.; Gueimonde, M. Characterization and in vitro properties of potentially probiotic bifidobacterium strains isolated from breast-milk. Int. J. Food Microbiol. 2011, 149, 28–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, A.; Farver, M.; Smilowitz, J. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr. Metab. Insights 2015, 8 (Suppl. S1), 1–9. [Google Scholar] [PubMed]
- Grguric, J.; Percl, M.; Kolacek, S.; Bacic, V. Microflora in the digestive tract of infants. Mljekarstvo 1996, 46, 291–296. [Google Scholar]
- Saavedra, J.M. Use of probiotics in pediatrics: Rationale, mechanisms of action, and practical aspects. Nutr. Clin. Pract. 2007, 22, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.T.; Totten, S.M.; Smilowitz, J.T.; Popovic, M.; Parker, E.; Lemay, D.G.; van Tassell, M.L.; Miller, M.J.; Jin, Y.S.; German, J.B.; et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, H.J.; Wildeboer–Veloo, A.C.; Raangs, G.C.; Wagendorp, A.A.; Klijn, N.; Bindels, J.G.; Welling, G.W. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 2000, 30, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Aggett, P.J.; Agostoni, C.; Axelsson, I.; Edwards, C.A.; Goulet, O.; Hernell, O.; Koletzko, B.; Lafeber, H.N.; Micheli, J.L.; Michaelsen, K.F.; et al. Nondigestible carbohydrates in the diets of infants and young children: A commentary by the ESPGHAN Committee on Nutrition. J. Pediatr. Gastroenterol. Nutr. 2003, 36, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Peano, C.; Pass, D.; Foroni, E.; Severgnini, M.; Claesson, M.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS ONE 2012, 7, e36957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turroni, F.; Duranti, S.; Bottacini, F.; Guglielmetti, S.; van Sinderen, D.; Ventura, M. Bifidobacterium bifidum as an example of a specialized human gut commensal. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Guglielmetti, S.; Mora, D.; Gschwender, M.; Popp, K. Randomised clinical trial: Bifidobacterium bifidum MIMBb75 significantly alleviates irritable bowel syndrome and improves quality of life—A double-BLIND, Placebo-Controlled Study. Aliment. Pharmacol. Ther. 2011, 33, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, Y. Effect of Lactobacillus acidophilus and Bifidobacterium bifidum supplementation to standard triple therapy on Helicobacter pylori eradication and dynamic changes in intestinal flora. World J. Microbiol. Biotechnol. 2013, 30, 847–853. [Google Scholar] [CrossRef] [PubMed]
- Toiviainen, A.; Jalasvuori, H.; Lahti, E.; Gursoy, U.; Salminen, S.; Fontana, M.; Flannagan, S.; Eckert, G.; Kokaras, A.; Paster, B.; et al. Impact of orally administered lozenges with lactobacillus rhamnosus GG and bifidobacterium animalis subsp. Lactis BB-12 on the number of salivary mutans streptococci, amount of plaque, gingival inflammation and the oral microbiome in healthy adults. Clin. Oral. Investig. 2014, 19, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Culpepper, T.; Christman, M.; Nieves, C.; Specht, G.; Rowe, C.; Spaiser, S.; Ford, A.; Dahl, W.; Girard, S.; Langkamp-Henken, B. Bifidobacterium bifidum R0071 decreases stress-associated diarrhoea-related symptoms and self-reported stress: A secondary analysis of a randomised trial. Benef. Microbes 2016, 7, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Kim, C.J.; Park, S.Y.; Ko, Y.T.; Jeong, H.K.; Ji, G.E. Growth and β-Glucosidase activity of bifidobacterium. J. Microbiol. Biotechnol. 1996, 6, 255–259. [Google Scholar]
- BIFIDO. Available online: http://www.bifido.com/en/product/probiotics/health?category=zigunuk (accessed on 30 June 2016).
- Kim, I.H.; Park, M.S.; Ji, G.E. Characterization of adhesion of Bifidobacterium sp. BGN4 to human enterocyte-like Caco-2 cells. J. Microbiol. Biotechnol. 2003, 13, 276–281. [Google Scholar]
- Ku, S.; You, H.J.; Ji, G.E. Enhancement of anti-tumorigenic polysaccharide production, adhesion, and branch formation of Bifidobacterium bifidum BGN4 by phytic acid. Food Sci. Biotechnol. 2009, 18, 749–754. [Google Scholar]
- Lee, M.J.; Zang, Z.; Choi, E.Y.; Shin, H.K.; Ji, G.E. Cytoskeleton reorganization and cytokine production of macrophages by bifidobacterial cells and cell-free extracts. J. Microbiol. Biotechnol. 2002, 12, 398–405. [Google Scholar]
- Kim, N.; Ji, G.E. Modulatory activity of Bifidobacterium sp. BGN4 cell fractions on immune cells. J. Microbiol. Biotechnol. 2006, 16, 584–589. [Google Scholar]
- Lee, S.; Koo, N.; Oh, S. Regulatory effect on specific ige response of Bifidobacterium bifidum (BGN4 Strain) in murine model of peanut allergy. J. Allergy Clin. Immunol. 2006, 117, S204. [Google Scholar] [CrossRef]
- Kim, N.; Kunisawa, J.; Kweon, M.; Ji, G.E.; Kiyono, H. Oral feeding of Bifidobacterium bifidum (BGN4) Prevents CD4 + CD45RB high T cell-mediated inflammatory bowel disease by inhibition of disordered T cell activation. Clin. Immunol. 2007, 123, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.S.; Kang, H.W.; Im, J.P.; Ji, G.E.; Kim, S.G.; Jung, H.C.; Song, I.S.; Kim, J.S. Effect of probiotics on symptoms in Korean adults with irritable bowel syndrome. Gut Liver 2009, 3, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kwon, J.H.; Ahn, S.H.; Lee, S.I.; Han, Y.S.; Choi, Y.O.; Lee, S.Y.; Ahn, K.M.; Ji, G.E. Effect of probiotic mix (Bifidobacterium bifidum, Bifidobacterium lactis, Lactobacillus acidophilus) in the primary prevention of eczema: A double-blind, randomized, placebo-controlled trial. Pediatr. Allergy Immunol. 2010, 21, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Park, M.S.; Ji, G.E. Probiotic modulation of dendritic cells co-cultured with intestinal epithelial cells. World J. Gastroenterol. 2012, 18, 1308–1318. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.Y.; Kim, Y.K.; Ji, G.E.; Om, A.S. Effect of fermented soymilk using Bifidobacterium spp. RD65 and BGN4 on abberant crypt foci in azoxymethane induced colon cancer rats. KoSFoST Int. Symp. Annu. Meet. 2001, 197. [Google Scholar]
- You, H.J.; Oh, D.K.; Ji, G.E. Anticancerogenic effect of a novel chiroinositol-containing polysaccharide from Bifidobacterium bifidum BGN4. FEMS Microbiol. Lett. 2004, 240, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Youn, S.Y.; Park, M.S.; Ji, G.E. Identification of the β-glucosidase gene from Bifidobacterium animalis Subsp. Lactis and its expression in B. bifidum BGN4. J. Microbiol. Biotechnol. 2012, 22, 1714–1723. [Google Scholar]
- Kim, J.Y.; Wang, Y.; Park, S.J.; Ji, G.E.; Park, M.S. Cloning and expression of β-Glucosidases from Bifidobacterium lactis AD011. Food Sci. Biotechnol. 2012, 21, 731–738. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, J.Y.; Park, M.S.; Ji, G.E. Novel Bifidobacterium promoters selected through microarray analysis lead to constitutive high-level gene expression. J. Microbiol. 2012, 50, 638–643. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.A.; Park, M.S.; Kang, S.A.; Ji, G.E. Production of γ-aminobutyric acid during fermentation of Gastrodia Elata Bl. By co-culture of Lactobacillus brevis GABA 100 with Bifidobacterium bifidum BGN4. Food Sci. Biotechnol. 2014, 23, 459–466. [Google Scholar] [CrossRef]
- You, H.J.; Ahn, H.J.; Kim, J.Y.; Wu, Q.Q.; Ji, G.E. High expression of β-glucosidase in Bifidobacterium bifidum BGN4 and application in conversion of isoflavone glucosides during fermentation of soy milk. J. Microbiol. Biotechnol. 2015, 25, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Seo, H.S.; Seo, J.M.; Suh, J.W.; Hwang, I.; Ji, G.E. Development of S-adenosyl-l-methionine (SAM)-reinforced probiotic yogurt using Bifidobacterium bifidum BGN4. Food Sci. Biotechnol. 2008, 17, 1025–1031. [Google Scholar]
- Kim, J.Y.; Suh, J.W.; Ji, G.E. Evaluation of S-adenosyl-l-methionine production by Bifidobacterium bifidum BGN4. Food Sci. Biotechnol. 2008, 17, 184–187. [Google Scholar]
- Yu, D.S.; Jeong, H.; Lee, D.H.; Kwon, S.K.; Song, J.Y.; Kim, B.Y.; Park, M.S.; Ji, G.E.; Oh, T.K.; Kim, J.F. Complete genome sequence of the probiotic bacterium Bifidobacterium bifidum strain BGN4. J. Bacteriol. 2012, 194, 4757–4758. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration, Bad Bug Book (Second Edition). Available online: http://www.fda.gov/Food/FoodborneIllnessContaminants/CausesOfIllnessBadBugBook/ (accessed on 30 June 2016).
- Ribet, D.; Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Thöle, C.; Brandt, S.; Ahmed, N.; Hensel, A. Acetylated rhamnogalacturonans from immature fruits of Abelmoschus esculentus inhibit the adhesion of Helicobacter pylori to human gastric cells by interaction with outer membrane proteins. Molecules 2015, 20, 16770–16787. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Shin, E.C.; Park, H.G. Fructooligosaccharides decreased the ability of probiotic Escherichia coli Nissle 1917 to adhere to co-cultures of human intestinal cell lines. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 45–52. [Google Scholar] [CrossRef]
- Serafini, F.; Strati, F.; Ruas-Madiedo, P.; Turroni, F.; Foroni, E.; Duranti, S.; Milano, F.; Perotti, A.; Viappiani, A.; Guglielmetti, S.; et al. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 2013, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Underwood, M.; Kananurak, A.; Coursodon, C.; Adkins-Reick, C.; Chu, H.; Bennett, S.; Wehkamp, J.; Castillo, P.; Leonard, B.; Tancredi, D.; et al. Bifidobacterium bifidum in a rat model of necrotizing enterocolitis: Antimicrobial peptide and protein responses. Pediatr. Res. 2012, 71, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.A.; Balciunas, E.M.; Converti, A.; Cotter, P.D.; de Souza Oliveira, R.P. Bacteriocin production by Bifidobacterium spp: A review. Biotechnol. Adv. 2013, 31, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Silva, O.N.; Franco, O.L. Recombinant probiotics with antimicrobial peptides: A dual strategy to improve immune response in immunocompromised patients. Drug Discov. Today 2014, 19, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Slavin, J. Fiber and prebiotics: Mechanisms and health benefits. Nutrients 2013, 5, 1417–1435. [Google Scholar] [CrossRef] [PubMed]
- Aronsson, L.; Huang, Y.; Parini, P.; Korach-André, M.; Håkansson, J.; Gustafsson, J.; Pettersson, S.; Arulampalam, V.; Rafter, J. Decreased fat storage by Lactobacillus paracasei is associated with increased levels of angiopoietin-like 4 protein (ANGPTL4). PLoS ONE 2010, 5, e13087. [Google Scholar] [CrossRef] [PubMed]
- DiBaise, J.K.; Frank, D.N.; Mathur, R. Impact of the gut microbiota on the development of obesity: Current concepts. Am. J. Gastroenterol. Suppl. 2012, 1, 22–27. [Google Scholar] [CrossRef]
- Hardy, H.; Harris, J.; Lyon, E.; Beal, J.; Foey, A. Probiotics, prebiotics and immunomodulation of gut mucosal defences: Homeostasis and immunopathology. Nutrients 2013, 5, 1869–1912. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Wang, Z.; Yang, Q. Ability of Lactobacillus to inhibit enteric pathogenic bacteria adhesion on Caco-2 cells. World J. Microbiol. Biotechnol. 2010, 27, 881–886. [Google Scholar] [CrossRef]
- Kim, B.J.; Hong, J.H.; Jeong, Y.S.; Jung, H.K. Evaluation of two Bacillus subtilis strains isolated from Korean fermented food as probiotics against loperamide-induced constipation in mice. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 797–806. [Google Scholar] [CrossRef]
- Lim, S.M. Anti-helicobacter pylori activity of antimicrobial substances produced by lactic acid bacteria isolated from Baikkimchi. J. Korean Soc. Appl. Biol. Chem. 2014, 57, 621–630. [Google Scholar] [CrossRef]
- Lebeer, S.; Vanderleyden, J.; De Keersmaecker, S.C. Host interactions of probiotic bacterial surface molecules: Comparison with commensals and pathogens. Nat. Rev. Microbiol. 2010, 8, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Van Tassell, M.; Miller, M. Lactobacillus adhesion to mucus. Nutrients 2011, 3, 613–636. [Google Scholar] [PubMed]
- Duary, R.K.; Rajput, Y.S.; Batish, V.K.; Grover, S. Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J. Med. Res. 2011, 134, 664–671. [Google Scholar] [PubMed]
- Polak-Berecka, M.; Waśko, A.; Paduch, R.; Skrzypek, T.; Sroka-Bartnicka, A. The effect of cell surface components on adhesion ability of Lactobacillus rhamnosus. Antonie van Leeuwenhoek 2014, 106, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Krasowska, A.; Sigler, K. How microorganisms use hydrophobicity and what does this mean for human needs? Front. Cell. Infect. Microbiol. 2014, 4, 112. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, C.; Bouley, C.; Cayuela, C.; Bouttier, S.; Bourlioux, P.; Bellon-Fontaine, M. Cell surface characteristics of Lactobacillus casei subsp. casei, Lactobacillus paracasei subsp. paracasei, and Lactobacillus rhamnosus strains. Appl. Environ. Microbiol. 1997, 63, 1725–1731. [Google Scholar] [PubMed]
- Abdulla, A.A.; Abed, T.A.; Saeed, A.M. Adhesion, Autoaggregation and hydrophobicity of six Lactobacillus strains. Br. Microbiol. Res. J. 2014, 4, 381–391. [Google Scholar] [CrossRef]
- Boris, S.; Suárez, J.E.; Vázquez, F.; Barbés, C. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect. Immun. 1998, 66, 1985–1989. [Google Scholar] [PubMed]
- Shakirova, L.; Auzina, L.; Zikmanis, P.; Gavare, M.; Grube, M. Influence of growth conditions on hydrophobicity of Lactobacillus acidophilus and Bifidobacterium lactis cells and characteristics by FT-IR spectra. J. Spectrosc. 2010, 24, 251–255. [Google Scholar] [CrossRef]
- Pérez, P.F.; Minnaard, Y.; Disalvo, E.A.; de Antoni, G.L. Surface properties of bifidobacterial strains of human origin. J. Microbiol. Biotechnol. 1998, 64, 21–26. [Google Scholar]
- Pan, W.; Li, P.; Liu, Z. The correlation between surface hydrophobicity and adherence of Bifidobacterium strains from centenarians’ faeces. Anaerobe 2006, 12, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Schillinger, U.; Guigas, C.; Holzapfel, W.H. In vitro adherence and other properties of lactobacilliused in probiotic yoghurt-like products. Int. Dairy J. 2005, 15, 1289–1297. [Google Scholar] [CrossRef]
- Alzate, A.; Fernandez, A.; Perez-Conde, M.C.; Gutierrez, A.M.; Camara, C. Comparison of biotransformation of inorganic selenium by Lactobacillus and Saccharomyces in lactic fermentation process of yogurt and kefir. J. Agric. Food Chem. 2008, 56, 8728–8736. [Google Scholar] [CrossRef] [PubMed]
- Botes, M.; Loos, B.; van Reenen, C.A.; Dicks, L.M.T. Adhesion of the probiotic strains enterococcus mundtii ST4SA and Lactobacillus plantarum 423 to Caco-2 cells under conditions simulating the intestinal tract, and in the presence of antibiotics and anti-inflammatory medicaments. Arch. Microbiol. 2008, 190, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Wadström, T.; Andersson, K.; Sydow, M.; Axelsson, L.; Lindgren, S.; Gullmar, B. Surface properties of lactobacilli isolated from the small intestine of pigs. J. Appl. Bacteriol. 1987, 62, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Candela, M.; Perna, F.; Carnevali, P.; Vitali, B.; Ciati, R.; Gionchetti, P.; Rizzello, F.; Campieri, M.; Brigidi, P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008, 125, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Crociani, J.; Grill, J.P.; Huppert, M.; Ballongue, J. Adhesion of different bifidobacteria strains to human enterocyte-like Caco-2 cells and comparison with in vivo study. Lett. Appl. Microbiol. 1995, 21, 146–148. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.T.; Selmi, C.; Meyers, F.J.; Keen, C.L.; Gershwin, M.E. Probiotics and immunity. J. Gastroenterol. 2009, 44, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Galdeano, C.M.; de LeBlanc, A.D.; Vinderola, G.; Bonet, M.B.; Perdigon, G. Proposed model: Mechanisms of immunomodulation induced by probiotic bacteria. Clin. Vaccine Immunol. 2007, 14, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.; Gordon, S. The M1 and M2 paradigm of macrophage activation: Time for reassessment. F1000Prime Rep. 2014, 6. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.I.; Joo, Y.H.; Pak, P.J.; Kim, J.S.; Chung, N. Different shapes of Al2O3 particles induce differential cytotoxicity via a mechanism involving lysosomal destabilization and reactive oxygen species generation. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 433–442. [Google Scholar] [CrossRef]
- Li, X.; Bao, W.; Leung, C.; Ma, D.; Zhang, G.; Lu, A.; Wang, S.; Han, Q. Chemical structure and immunomodulating activities of an α-glucan purified from Lobelia chinensis lour. Molecules 2016, 21, 779. [Google Scholar] [CrossRef] [PubMed]
- Cui, S.; Hassan, R.; Heintz-Buschart, A.; Bilitewski, U. Regulation of Candida albicans interaction with macrophages through the activation of HOG pathway by genistein. Molecules 2016, 21, 162. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.S.; Rutherfurd, K.J.; Cross, M.L.; Gopal, P.K. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 2001, 74, 833–839. [Google Scholar] [PubMed]
- Fong, F.L.; Shah, N.P.; Kirjavainen, P.; El-Nezami, H. Mechanism of action of probiotic bacteria on intestinal and systemic immunities and antigen-presenting cells. Int. Rev. Immunol. 2015, 1–11. [Google Scholar] [CrossRef] [PubMed]
- World Cancer Research Fund International, Colorectal Cancer Statistics. Available online: http://www.wcrf.org/int/cancer-facts-figures/data-specific-cancers/colorectal-cancer-statistics (accessed on 5 July 2016).
- American Cancer Society, Key Statistics for Colorectal Cancer. Available online: http://www.cancer.org/cancer/colonandrectumcancer/detailedguide/colorectal-cancer-key-statistics (accessed on 12 July 2016).
- Pourhoseingholi, M. Increased burden of colorectal cancer in Asia. World J. Gastrointest. Oncol. 2012, 4, 68–70. [Google Scholar] [CrossRef] [PubMed]
- Selhub, E.; Logan, A.; Bested, A. Fermented foods, microbiota, and mental health: Ancient practice meets nutritional psychiatry. J. Physiol. Anthropol. 2014, 33. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Nagpal, R.; Verma, V.; Kumar, A.; Kaur, N.; Hemalatha, R.; Gautam, S.; Singh, B. Probiotic metabolites as epigenetic targets in the prevention of colon cancer. Nutr. Rev. 2012, 71, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.S.; Kim, Y.; Han, K.S.; You, S.; Oh, S.; Kim, S.H. Effects of Lactobacillus strains on cancer cell proliferation and oxidative stress in vitro. Lett. Appl. Microbiol. 2006, 42, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Uccello, M.; Malaguarnera, G.; Basile, F.; D’agata, V.; Malaguarnera, M.; Bertino, G.; Vacante, M.; Drago, F.; Biondi, A. Potential role of probiotics on colorectal cancer prevention. BMC Surg. 2012, 12, S35. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Ambalam, P.; Doble, M. Probiotics and Bioactive Carbohydrates in Colon Cancer Management; Springer (India) Pvt. Ltd.: New Delhi, India, 2016; pp. 83–109. [Google Scholar]
- Sadeghi-Aliabadi, H.; Mohammadi, F.; Fazeli, H.; Mirlohi, M. Effects of Lactobacillus plantarum A7 with probiotic potential on colon cancer and normal cells proliferation in comparison with a commercial strain. Iran J. Basic Med. Sci. 2014, 17, 815–819. [Google Scholar] [PubMed]
- Nagaoka, M.; Hashimoto, S.; Watanabe, T.; Yokokura, T.; Mori, Y. Anti-ulcer effects of lactic acid bacteria and their cell wall polysaccharides. Biol. Pharm. Bull. 1994, 17, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Van Breemen, R.; Li, Y. Caco-2 cell permeability assays to measure drug absorption. Expert Opin. Drug. Metab. Toxicol. 2005, 1, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Corpet, D.; Tache, S. Most effective colon cancer chemopreventive agents in rats: A systematic review of aberrant crypt foci and tumor data, ranked by potency. Nutr. Cancer 2002, 43, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Ng, J.; Arozulllah, A.; Ewing, R.; Llor, X.; Carroll, R.E.; Benya, R.V. Aberrant crypt focus size predicts distal polyp histopathology. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Foerst, P.; Santivarangkna, C. Advances in Probiotic Technology; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2015; pp. 356–374. [Google Scholar]
- Foligné, B.; Daniel, C.; Pot, B. Probiotics from research to market: The possibilities, risks and challenges. Curr. Opin. Microbiol. 2013, 16, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Lahtinen, S. Probiotic viability—Does it matter? Microb. Ecol. Health Dis. 2012, 23, 10. [Google Scholar] [CrossRef] [PubMed]
- Nedović, V.; Raspor, P.; Lević, J.; Tumbas Šaponjac, V.; Barbosa-Cánovas, G. Emerging and Traditional Technologies for Safe, Healthy and Quality Food; Springer International Publishing: Cham, Germany, 2016; pp. 257–268. [Google Scholar]
- Park, S.J.; Youn, S.Y.; Ji, G.E.; Park, M.S. Whole cell biotransformation of major ginsenosides using leuconostocs and lactobacilli. Food Sci. Biotechnol. 2012, 21, 839–844. [Google Scholar] [CrossRef]
- You, H.J.; Ahn, H.J.; Ji, G.E. Transformation of Rutin to antiproliferative quercetin-3-glucoside by Aspergillus niger. J. Agric. Food Chem. 2010, 58, 10886–10892. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; Zheng, H.; Park, M.S.; Ji, G.E. Optimization of β-glucuronidase activity from Lactobacillus delbrueckii Rh2 and its use for biotransformation of baicalin and wogonoside. J. Korean Soc. Appl. Biol. Chem. 2011, 54, 275–280. [Google Scholar] [CrossRef]
- Ku, S.; You, H.J.; Park, M.S.; Ji, G.E. Effects of ascorbic acid on α-l-arabinofuranosidase and α-l-arabinopyranosidase activities from Bifidobacterium longum RD47 and its application to whole cell bioconversion of ginsenoside. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Ku, S.; You, H.J.; Park, M.S.; Ji, G.E. Whole-cell biocatalysis for producing ginsenoside Rd from Rb1 using Lactobacillus rhamnosus GG. J. Microbiol. Biotechnol. 2016, 26, 1206–1215. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Zhang, J.M.; Wang, Z.G.; Peng, W.; Hu, H.L.; Fu, C.M. Biotransformation, a promising technology for anti-cancer drug development. Asian Pac. J. Cancer Prev. 2013, 14, 5599–5608. [Google Scholar] [CrossRef] [PubMed]
- Sasidharan, S.; Chen, Y.; Saravanan, D.; Sundram, K.; Latha, L. Extraction, isolation and characterization of bioactive compounds from plants’ extracts. Afr. J. Tradit. Complement Altern. Med. 2011, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Han, B.H.; Park, M.H.; Han, Y.N.; Woo, L.K.; Sankawa, U.; Yahara, S.; Tanaka, O. Degradation of ginseng saponins under mild acidic conditions. Planta Med. 1982, 44, 146–149. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Nose, M.; Ogihara, Y. Alkaline cleavage of ginsenosides. Chem. Pharm. Bull. 1987, 35, 1653–1655. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.A.; Qin, J.J.; Wang, W.; Wang, M.H.; Wang, H.; Zhang, R. Ginsenosides as anticancer agents: In vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front. Pharmacol. 2012, 3. [Google Scholar] [CrossRef] [PubMed]
- Taku, K.; Melby, M.; Nishi, N.; Omori, T.; Kurzer, M. Soy isoflavones for osteoporosis: An evidence-based approach. Maturitas 2011, 70, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Orgaard, A.; Jensen, L. The effects of soy isoflavones on obesity. Exp. Biol. Med. 2008, 233, 1066–1080. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wang, R.; Song, X.; Chibbar, R.; Wang, X.; Wu, L.; Meng, Q. Dietary soy isoflavones increase insulin secretion and prevent the development of diabetic cataracts in streptozotocin-induced diabetic rats. Nutr. Res. 2008, 28, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Rafii, F. The role of colonic bacteria in the metabolism of the natural isoflavone daidzin to equol. Metabolites 2015, 5, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, A.R. The evidence for soybean products as cancer preventive agents. J. Nutr. 1995, 125, 733–743. [Google Scholar]
- Bawa, S. The significance of soy protein and soy bioactive compounds in the prophylaxis and treatment of osteoporosis. J. Osteoporos. 2010, 2010, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.; Wu, R.; Lee, K. A process for high-efficiency isoflavone deglycosylation using Bacillus subtilis natto NTU-18. Appl. Microbiol. Biotechnol. 2012, 94, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- New York Times, Stop Bashing G.M.O. Foods, More Than 100 Nobel Laureates Say. Available online: http://www.nytimes.com/2016/07/01/us/stop-bashing-gmo-foods-more-than-100-nobel-laureates-say.html?_r=0 (accessed on 12 July 2016).
- Hayakawa, K.; Kimura, M.; Kasaha, K.; Matsumoto, K.; Sansawa, H.; Yamori, Y. Effect of a γ-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br. J. Nutr. 2004, 92, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Adeghate, E.; Ponery, A. GABA in the endocrine pancreas: Cellular localization and function in normal and diabetic rats. Tissue Cell 2002, 34, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; Mazzacane, F.; Rizzello, C.; de Angelis, M.; Giuliani, G.; Meloni, M.; de Servi, B.; Gobbetti, M. Synthesis of γ-aminobutyric acid (GABA) by Lactobacillus plantarum DSM19463: Functional grape must beverage and dermatological applications. Appl. Microbiol. Biotechnol. 2009, 86, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qiu, T.; Huang, G.; Cao, Y. Production of γ-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb. Cell Fact. 2010, 9. [Google Scholar] [CrossRef] [PubMed]
- Huh, K.; Yi, S.J.; Shin, U.S.; Park, J.M. Effect of the ether fraction of Gastrodia elata methanol extract on the pentylenetetrazole-induced seizures. J. Appl. Pharmacol. 1995, 3, 199–204. [Google Scholar]
- Lee, O.H.; Kim, K.I.; Han, C.K.; Kim, Y.C.; Hong, H.D. Effects of acidic polysaccharides from Gastrodia rhizome on systolic blood pressure and serum lipid concentrations in spontaneously hypertensive rats fed a high-fat diet. Int. J. Mol. Sci. 2012, 13, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Lee, M.Y.; Ji, G.E.; Lee, Y.S.; Hwang, K.T. Production of γ-aminobutyric acid in black raspberry juice during fermentation by Lactobacillus brevis GABA100. Int. J. Food Microbiol. 2009, 130, 12–16. [Google Scholar] [CrossRef] [PubMed]
- Komatsuzaki, N.; Shima, J.; Kawamoto, S.; Momose, H.; Kimura, T. Production of γ-aminobutyric acid (gaba) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005, 22, 497–504. [Google Scholar] [CrossRef]
- Matos, J.R.; Raushel, F.; Wong, C.H. S-adenosylmethionine: Studies on chemical and enzymatic synthesis. Biotechnol. Appl. Biochem. 1987, 9, 39–52. [Google Scholar] [PubMed]
- Papakostas, G.; Alpert, J.; Fava, M. S-adenosyl-methionine in depression: A comprehensive review of the literature. Curr. Psychiatry Rep. 2003, 5, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Lieber, C.S. S-adenosyl-l-methionine: Its role in the treatment of liver disorders. Am. J. Clin. Nutr. 2002, 76, 1183–1187. [Google Scholar]
- Häuser, W.; Bernardy, K.; Üçeyler, N.; Sommer, C. Treatment of fibromyalgia syndrome with antidepressants. J. Gen. Intern. Med. 2009, 301, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Armando, M.; Galvagno, M.; Dogi, C.; Cerrutti, P.; Dalcero, A.; Cavaglieri, L. Statistical optimization of culture conditions for biomass production of probiotic gut-borne Saccharomyces cerevisiae strain able to reduce fumonisin B1. J. Appl. Microbiol. 2013, 114, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.; Appaiah, A. Optimization of culture conditions for bacterial cellulose production from Gluconacetobacter hansenii UAC09. Ann. Microbiol. 2011, 61, 781–787. [Google Scholar] [CrossRef]
- Kwon, S.G.; Son, J.W.; Kim, H.J.; Park, C.S.; Lee, J.K.; Ji, G.E.; Oh, D.K. High concentration cultivation of Bifidobacterium bifidum in a submerged membrane bioreactor. Biotechnol. Prog. 2006, 22, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Ji, G.E.; Ku, S.; Park, M.S. Culture medium Containing Phytic Acid for Cultivation of Bifidobacterium bifidum BGN4 and Method for Production of Bifidobacterium bifidum BGN4 Polysaccharide Using the Medium. S. Korea Patent 1010377780000, 23 May 2011. [Google Scholar]
- Dias, F.F.; Okrend, H.; Dondero, N.C. Calcium nutrition of Sphaerotilus growing in a continuous-flow apparatus. Appl. Microbiol. 1968, 16, 1364–1369. [Google Scholar] [PubMed]
- Snellen, J.E.; Raj, H.D. Morphogenesis and fine structure of Leucothrix mucor and effects of calcium deficiency. J. Bacteriol. 1970, 101, 240–249. [Google Scholar] [PubMed]
- Wright, C.T.; Klaenhammer, T.R. Calcium-induced alteration of cellular morphology affecting the resistance of Lactobacillus acidophilus to freezing. Appl. Environ. Microbiol. 1981, 41, 807–815. [Google Scholar] [PubMed]
- Apás, A.; Arena, M.; Colombo, S.; González, S. Probiotic administration modifies the milk fatty acid profile, intestinal morphology, and intestinal fatty acid profile of goats. J. Dairy Sci. 2015, 98, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Kojima, M.; Suda, S.; Hotta, S.; Hamada, K.; Suganuma, A. Necessity of calcium ion for cell division in Lactobacillus bifidus. J. Bacteriol. 1970, 104, 1010–1013. [Google Scholar] [PubMed]
- Duranti, S.; Milani, C.; Lugil, G.A.; Turroni, F.; Mancabelli, L.; Sanchez, B.; Ferrario, C.; Viappiani, A.; Mangifestra, M.; Mancino, W.; et al. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ. Microbiol. 2015, 17, 2515–2531. [Google Scholar] [CrossRef] [PubMed]
- Lugil, G.A.; Millani, C.; Turroni, F.; Duranti, S.; Ferrario, C.; Viappiani, A.; Mancabelli, L.; Mangifesta, M.; Taminiau, B.; Delcenserie, V.; et al. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl. Environ. Microbiol. 2014, 80, 6383–6394. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Turroni, F.; Duranti, S.; Lugil, G.A.; Mancabelli, L.; Ferrario, C.; Sinderen, D.; Ventura, M. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl. Environ. Microbiol. 2016, 82, 980–991. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; Canchaya, C.; Fitzgerald, G.F.; Gupta, R.S.; Sinderen, D. Genomics as a means to understand bacterial phylogeny and ecological adaptation: The case of bifidobacteria. Antonie van Leeuwenhoek 2007, 91, 351–372. [Google Scholar] [CrossRef] [PubMed]
- Ventura, M.; O’Connell-Motherway, M.; Leahy, S.; Moreno-Munoz, J.A.; Fitzgerald, G.F.; Sinderen, D. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Mirobiol. 2007, 120, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Bottacini, F.; Foroni, E.; Mulder, I.; Kim, J.H.; Zomer, A.; Sanchez, B.; Bidossi, A.; Ferrarini, A.; Giubellini, V.; et al. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc. Natl. Acad. Sci. USA 2010, 107, 19514–19519. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Milani, C.; Duranti, S.; Mancabelli, L.; Mangifestra, M.; Viappiani, A.; Lugil, G.A.; Ferrario, C.; Gioiosa, L.; Ferrarini, A.; et al. Deciphering bifidobacterial-mediaated metabolic interactions and their impact of gut microbiota by a multi-omics approach. ISME J. 2016, 10, 1656–1668. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.A.; Ross, R.P.; Fitzgerald, G.F.; Stanton, C. Metabolic activities and probiotic potential of bifidobacteria. Int. J. Food Microbiol. 2011, 149, 88–105. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Lugil, G.A.; Duranti, S.; Turroni, F.; Bottacini, F.; Mangifestra, M. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl. Environ. Microbiol. 2014, 80, 6290–6302. [Google Scholar] [CrossRef] [PubMed]
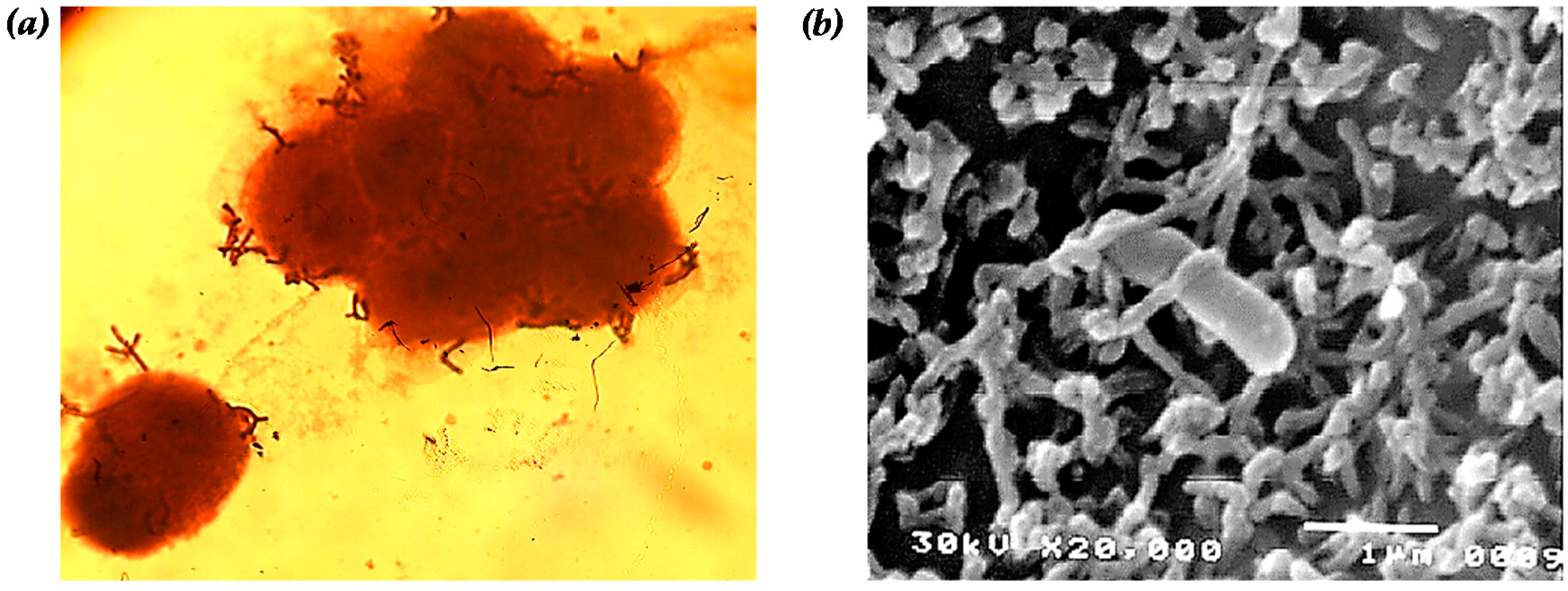
| No. | Cell | CHS (%) | No. | Cell | CHS (%) |
|---|---|---|---|---|---|
| 1 | B. bifidum BGN4 | 93 | 20 | B. longum ATCC 15707 | <5 |
| 2 | Bifidobacterium KJ | 90 | 21 | B. longums P-3 | 18.5 |
| 3 | Bifidobacterium HJ-30 | 90 | 22 | B. animalis H-9 | 37.13 |
| 4 | B. adolescentis ATCC 15703 | 90 | 23 | B. animalis P-4 | 17.4 |
| 5 | B. animalis ATCC 2552 | 86 | 24 | B. asteroids H-10 | 49.5 |
| 6 | B. animalis M6 | 85 | 25 | B. pseudocatenulatum I-6 | 47.3 |
| 7 | B. animalis Rd60 | 69.6 | 26 | B. pseudolongum CIDCA | 85 |
| 8 | B. animalis SI | 66.3 | 27 | B. lactis Bb12 | 75 |
| 9 | B. animalis CN2 | 21 | 28 | L. acidophilus LA5 | 75.1 |
| 10 | B. bifidum ATCC 2952 | 12 | 29 | L. paracasei (lac 1) | 80 |
| 11 | B. bifidum RD54 | 7 | 30 | L. acidophilus (lac 2) | 65 |
| 12 | B. bifidum MS1 | 6 | 31 | L. acidophilus (lac 3) | 60 |
| 13 | B. bifidum SH5 | 6 | 32 | L. acidophilus (lac 4) | 30 |
| 14 | B. bifidum E15 | 5 | 33 | L. fermentum (lac 5) | 45 |
| 15 | B. bifidum E2-18 | <5 | 34 | L. fermentum (lac 6) | 65 |
| 16 | B. bifidum JS9 | <5 | 35 | L. acidophilus | 80 |
| 17 | B. bifidum SH2 | <5 | 36 | L. gasseri | 80 |
| 18 | B. bifidum SJ32 | <5 | 37 | L. jensenii | 80 |
| 19 | B. infantis ATCC 15697 | <5 | - | - | - |
| Strain Name | B. bifidum BGN4 | B. bifidum PRL2010 | B. bifidum S17 |
|---|---|---|---|
| Accession | NC_017999.1 | NC_014638.1 | NC_014616.1 |
| Sequencing Status | Complete | Complete | Complete |
| Genome Size (bp) | 2,223,664 | 2,214,656 | 2,186,882 |
| G + C ratio (%) | 62.65 | 62.67 | 62.76 |
| Number of Chromosones | 1 | 1 | 1 |
| Number of Contigs | 1 | 1 | 1 |
| Number of ORFs | 1834 | 1706 | 1783 |
| Number of rRNA Genes | 9 | 9 | 9 |
| Number of tRNA Genes | 52 | 52 | 53 |
| COG | Description | B. bifidum BGN4 | B. bifidum PRL2010 | B. bifidum S17 | |||
|---|---|---|---|---|---|---|---|
| Number of Genes | % | Number of Genes | % | Number of Genes | % | ||
| J | Translation, ribosomal structure and biogenesis | 136 | 10.56% | 135 | 10.39% | 135 | 10.48% |
| K | Transcription | 95 | 7.38% | 95 | 7.31% | 93 | 7.22% |
| L | Replication, recombination and repair | 102 | 7.92% | 107 | 8.24% | 100 | 7.76% |
| D | Cell cycle control, cell division, chromosome partitioning | 24 | 1.86% | 22 | 1.69% | 23 | 1.79% |
| O | Posttranslational modification, protein turnover, chaperones | 50 | 3.88% | 50 | 3.85% | 50 | 3.88% |
| M | Cell wall/membrane/envelope biogenesis | 75 | 5.82% | 81 | 6.24% | 79 | 6.13% |
| N | Cell motility | 6 | 0.47% | 6 | 0.46% | 5 | 0.39% |
| P | Inorganic ion transport and metabolism | 50 | 3.88% | 49 | 3.77% | 49 | 3.80% |
| T | Signal transduction mechanisms | 47 | 3.65% | 50 | 3.85% | 47 | 3.65% |
| C | Energy production and conversion | 50 | 3.88% | 50 | 3.85% | 51 | 3.96% |
| G | Carbohydrate transport and metabolism | 118 | 9.16% | 117 | 9.01% | 118 | 9.16% |
| E | Amino acid transport and metabolism | 135 | 10.48% | 137 | 10.55% | 136 | 10.56% |
| F | Nucleotide transport and metabolism | 56 | 4.35% | 55 | 4.23% | 56 | 4.35% |
| H | Coenzyme transport and metabolism | 45 | 3.49% | 44 | 3.39% | 44 | 3.42% |
| I | Lipid transport and metabolism | 35 | 2.72% | 36 | 2.77% | 36 | 2.80% |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 6 | 0.47% | 7 | 0.54% | 6 | 0.47% |
| R | General function prediction only | 150 | 11.65% | 148 | 11.39% | 153 | 11.88% |
| S | Function unknown | 108 | 8.39% | 110 | 8.47% | 107 | 8.31% |
| Total | 1288 | 100% | 1299 | 100% | 1288 | 100% | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, S.; Park, M.S.; Ji, G.E.; You, H.J. Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. Int. J. Mol. Sci. 2016, 17, 1544. https://doi.org/10.3390/ijms17091544
Ku S, Park MS, Ji GE, You HJ. Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism. International Journal of Molecular Sciences. 2016; 17(9):1544. https://doi.org/10.3390/ijms17091544
Chicago/Turabian StyleKu, Seockmo, Myeong Soo Park, Geun Eog Ji, and Hyun Ju You. 2016. "Review on Bifidobacterium bifidum BGN4: Functionality and Nutraceutical Applications as a Probiotic Microorganism" International Journal of Molecular Sciences 17, no. 9: 1544. https://doi.org/10.3390/ijms17091544