Comprehensive Proteomic Analysis of Spider Dragline Silk from Black Widows: A Recipe to Build Synthetic Silk Fibers
Abstract
:1. Introduction
2. Results
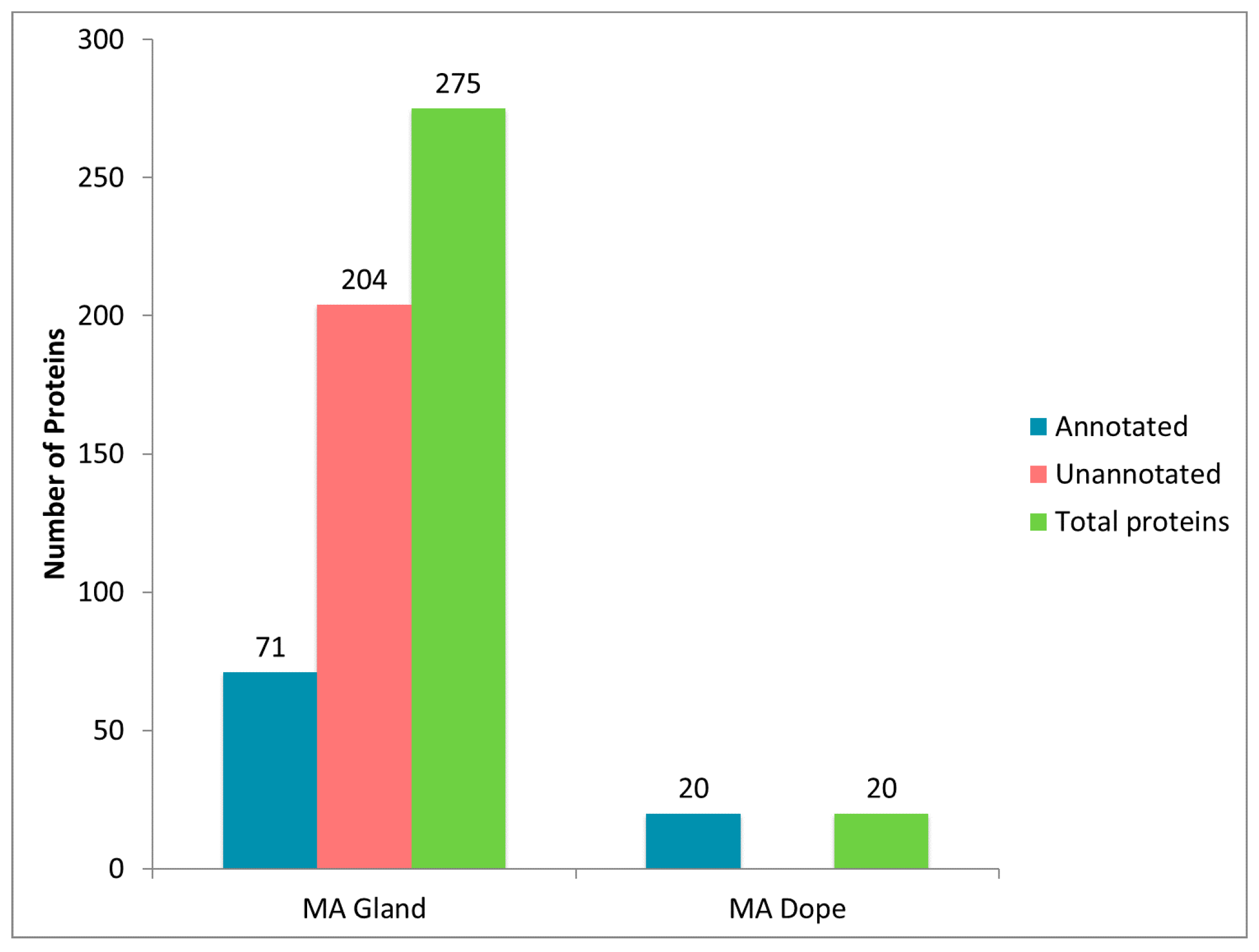
2.1. Diverse Chemical Solvent Treatments of MA Glands, Its Spinning Dope, and Dragline Silk Fibers Reveal Differential Protein Solubilization
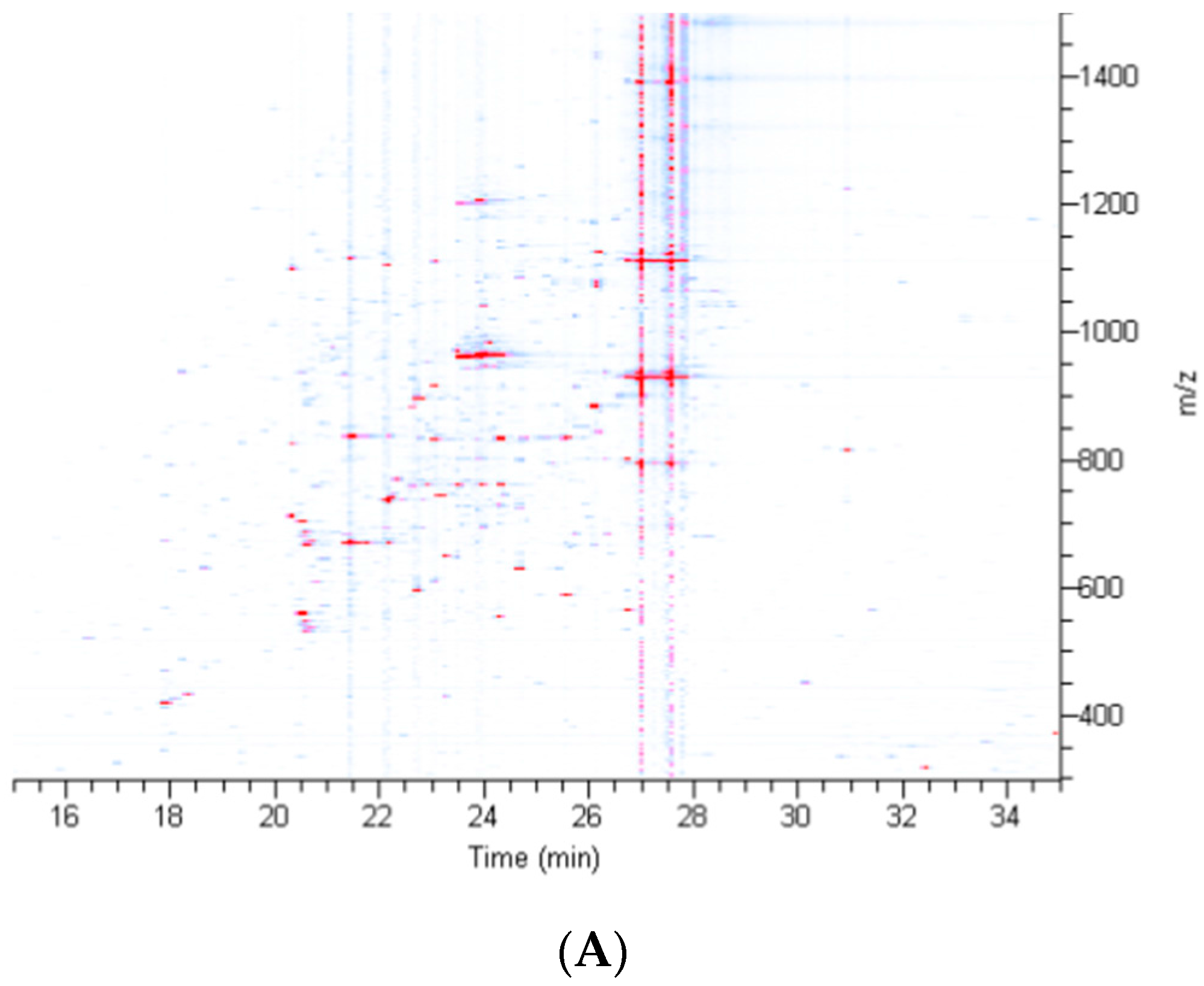
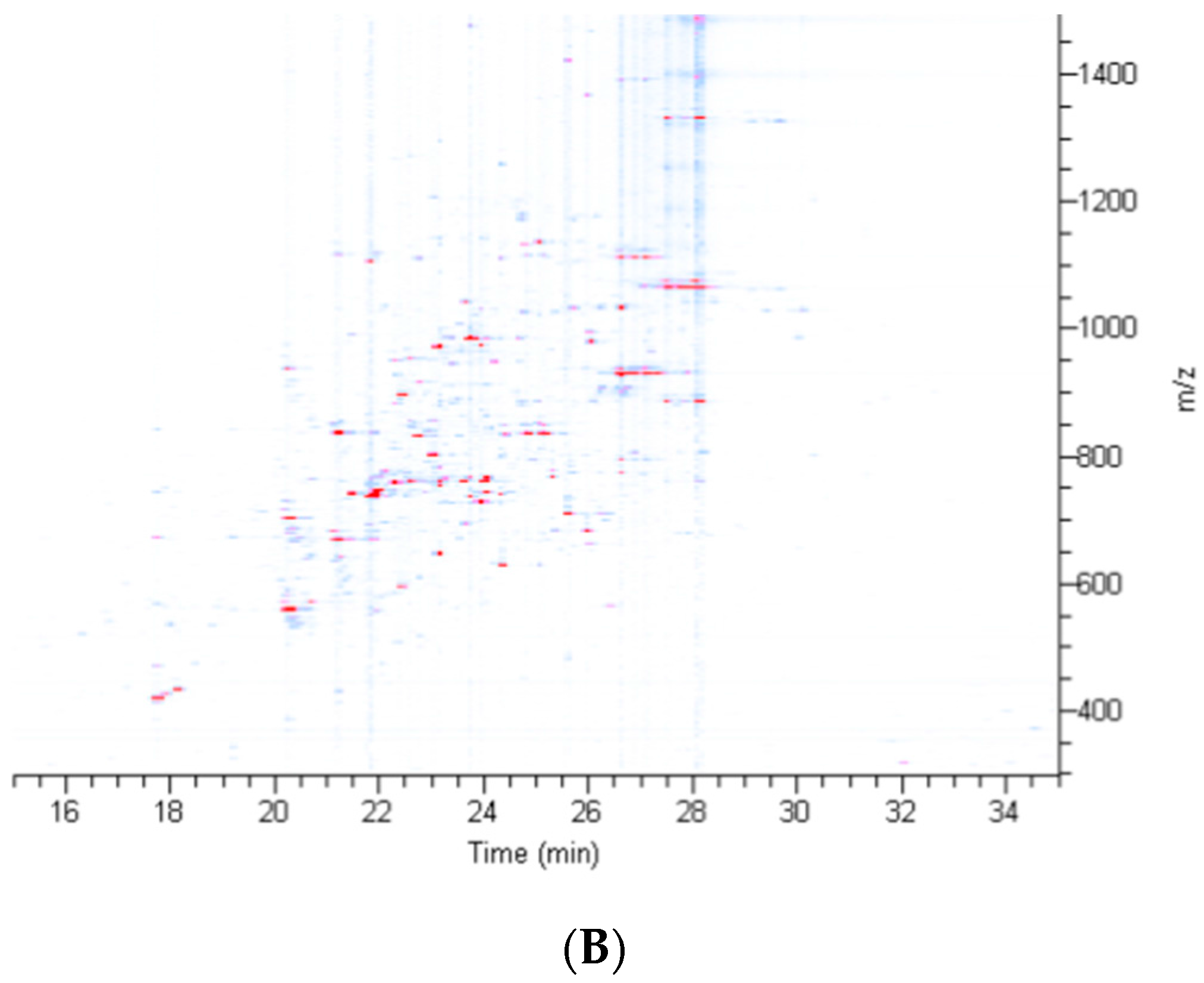
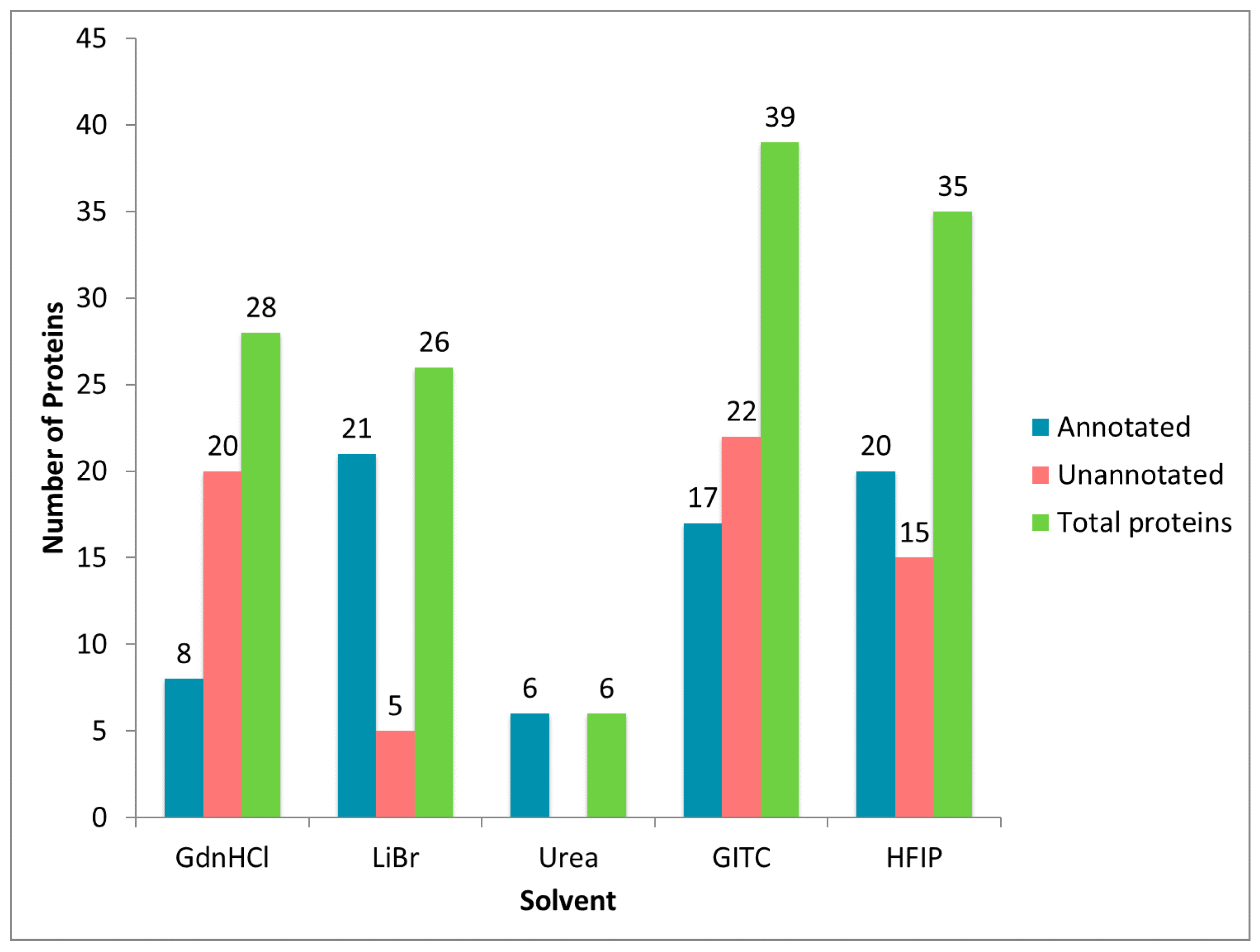
2.2. Treatment of Dragline Silk with GITC Leads to the Identification of the Highest Number of Proteins by MS/MS Analysis
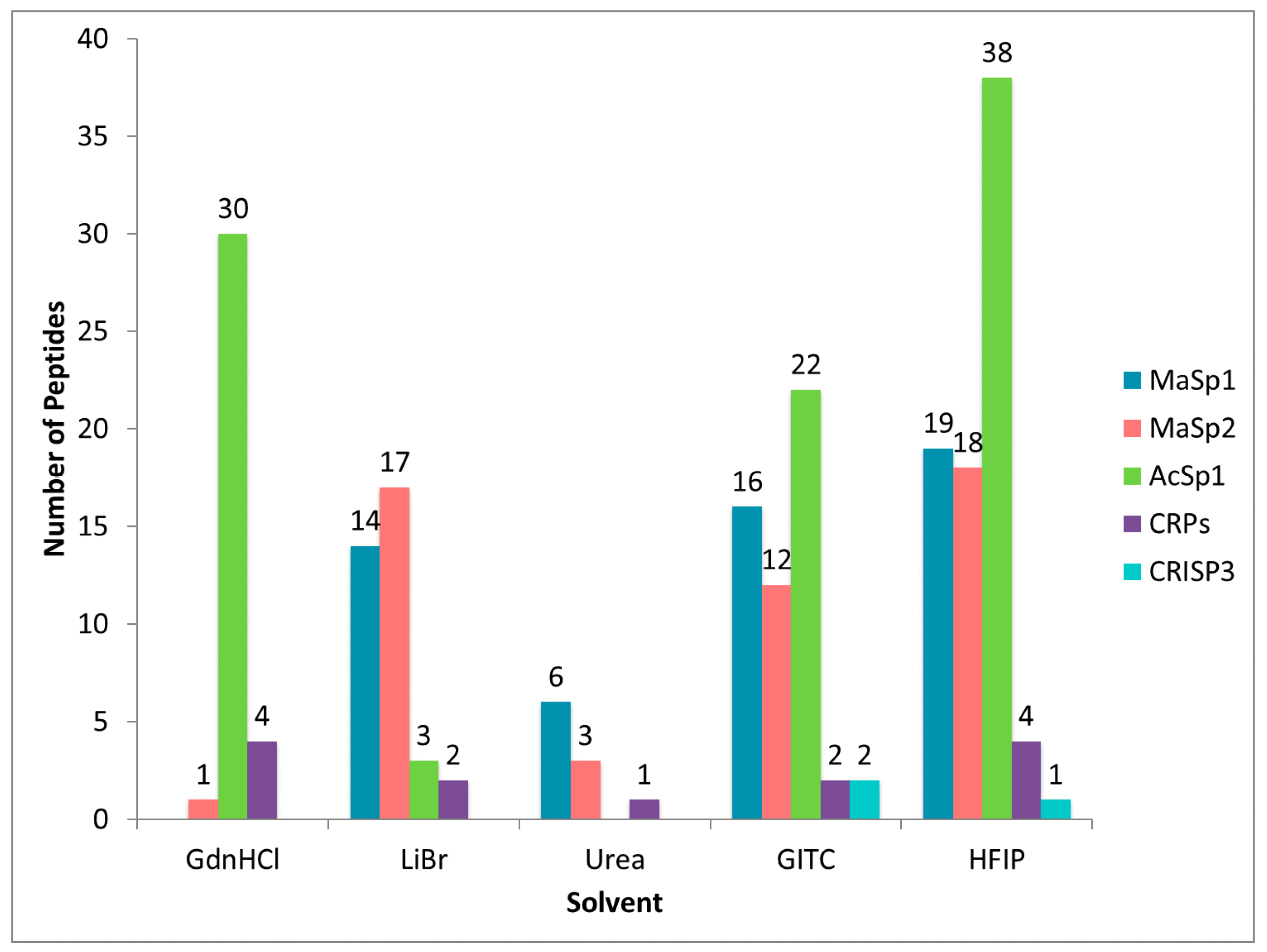
2.3. Different Solvent Systems Solubilize Dragline Silk Proteins with Different Efficiencies
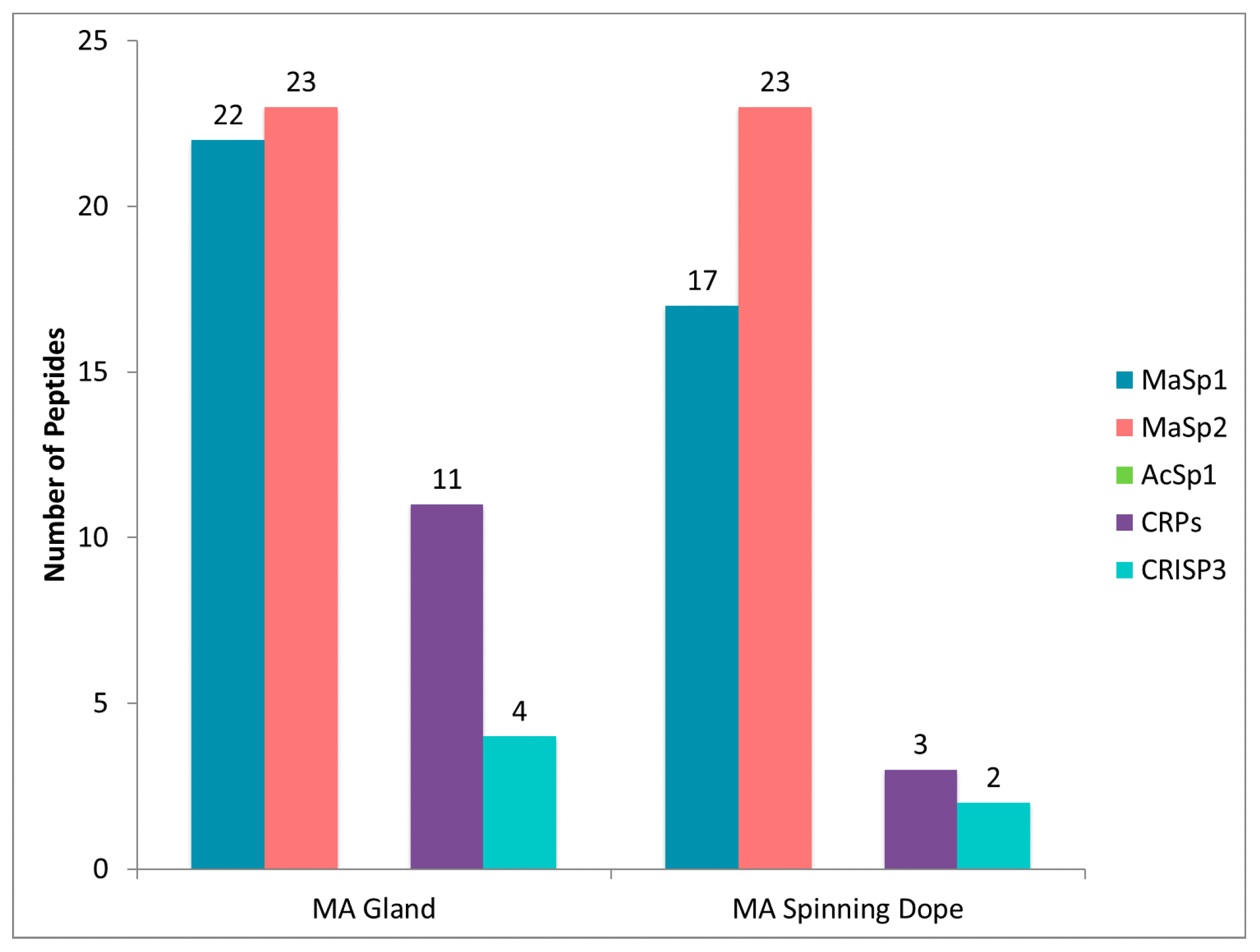
2.4. CRP1, CRP2 and CRP4 Are Present within MA Glands, Spinning Dope, and Dragline Silk Fibers
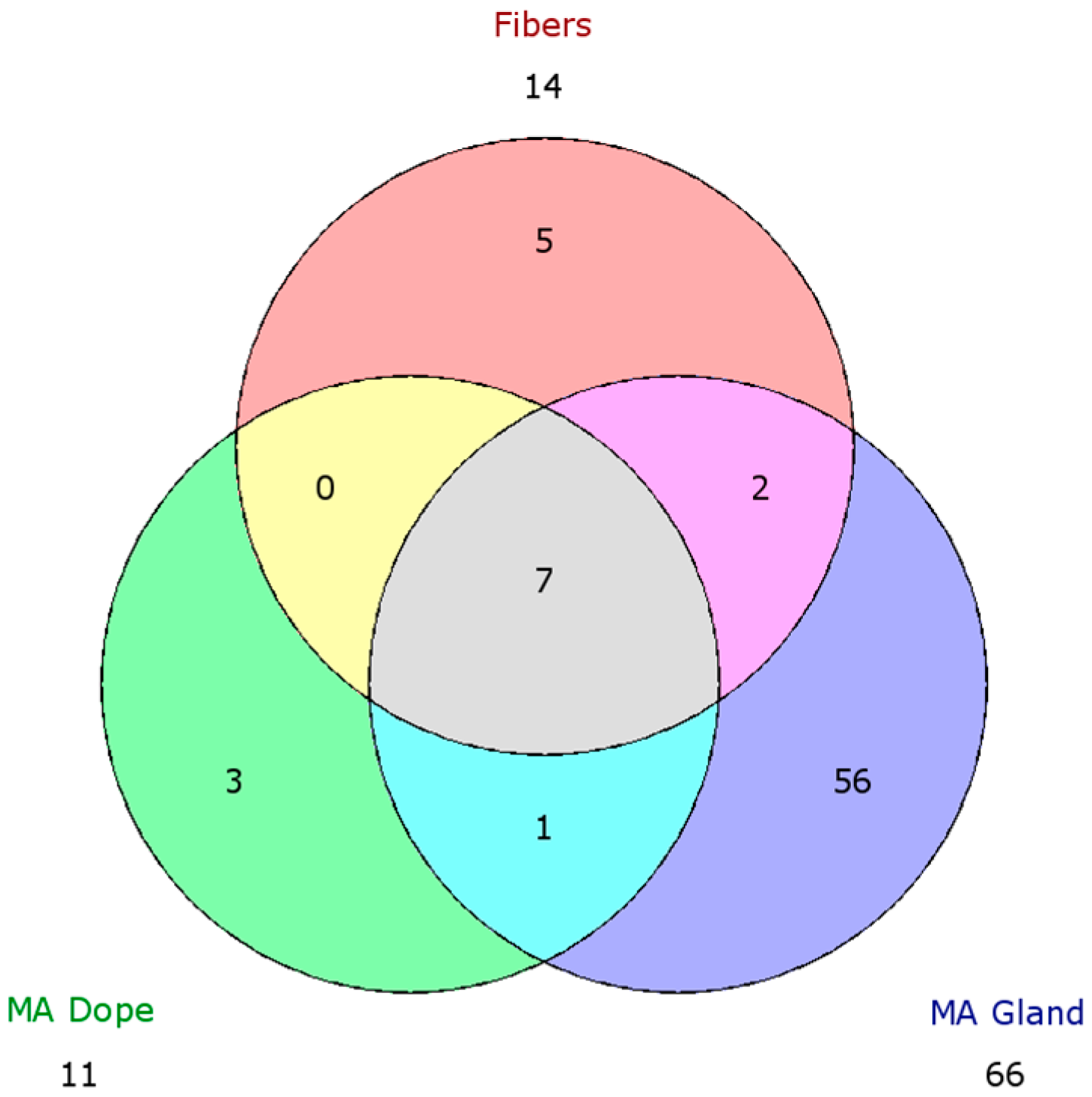
2.5. Comparison of Proteomic Data Sets
3. Discussion
3.1. Fundamental Knowledge of Silk Extrusion Pathway
3.2. Proteomic Analysis with Different Chemical Solvents
3.3. CRP1, CRP2 and CRP4 Are Present within Dragline Silk Fibers
3.4. New Paradigm Shift Is Emerging that Supports Different Silk Types Are Spun from Multiple Spidroin Family Members
4. Experimental Section
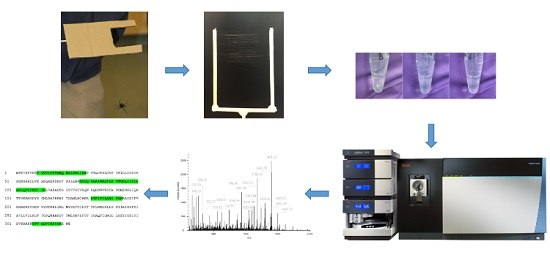
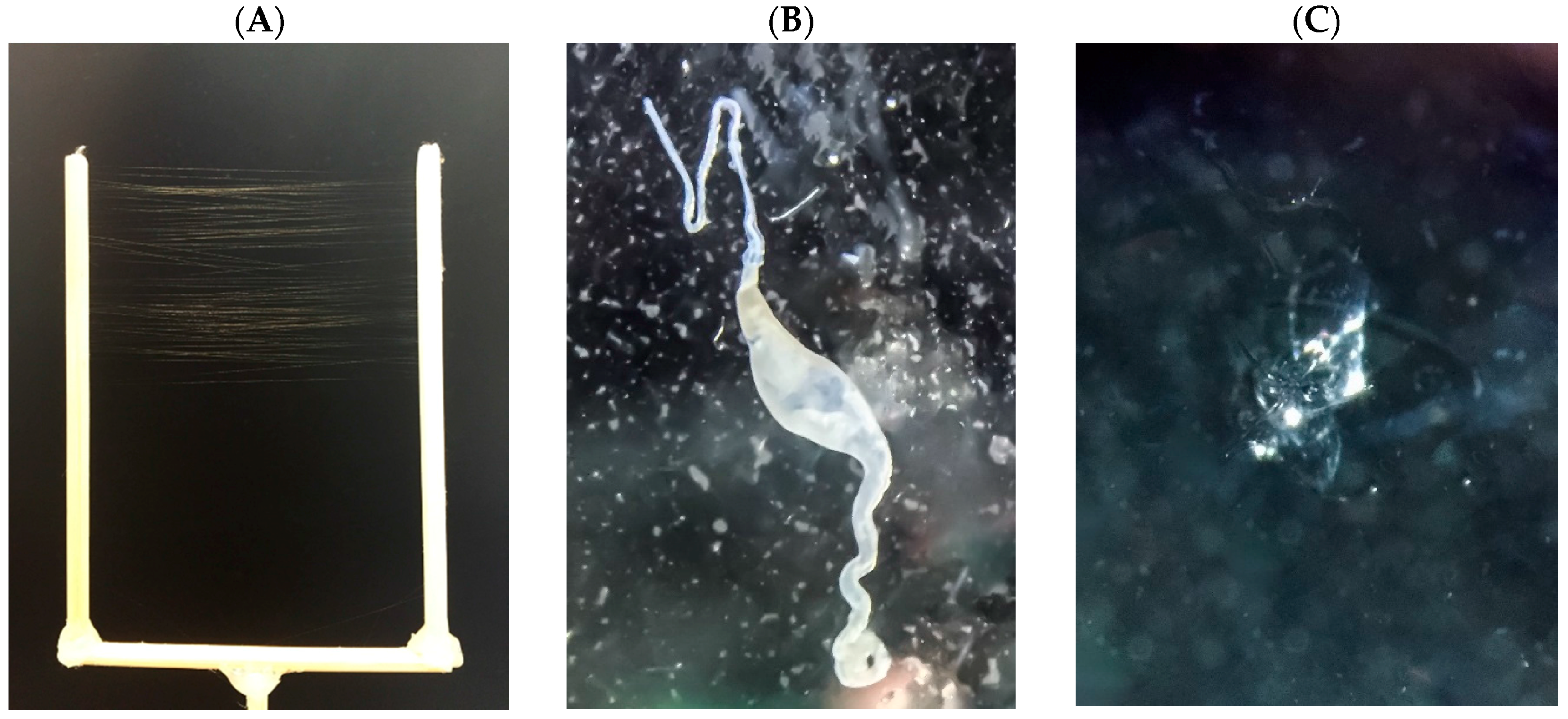
4.1. Silk Collection
4.2. Gland Dissection
4.3. Protein Extraction
4.4. Tryptic Digestion
4.5. Chromatography
4.6. Mass Spectrometry
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gosline, J.M.; DeMont, M.E.; Denny, M.W. The structure and properties of spider silk. Endeavour 1986, 10, 31–43. [Google Scholar] [CrossRef]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber: Spider dragline silk. Proc. Natl. Acad. Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.B.; Lewis, R.V. Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol. Chem. 1992, 267, 19320–19324. [Google Scholar]
- Teule, F.; Cooper, A.R.; Furin, W.A.; Bittencourt, D.; Rech, E.L.; Brooks, A.; Lewis, R.V. A protocol for the production of recombinant spider silk-like proteins for artificial fiber spinning. Nat. Protoc. 2009, 4, 341–355. [Google Scholar] [CrossRef] [PubMed]
- Hsia, Y.; Gnesa, E.; Pacheco, R.; Kohler, K.; Jeffery, F.; Vierra, C. Synthetic spider silk production on a laboratory scale. J. Vis. Exp. 2012. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, A.; Scheibel, T. Recombinant production of spider silk proteins. Adv. Appl. Microbiol. 2013, 82, 115–153. [Google Scholar] [PubMed]
- Anthoula Lazaris, S.A.; Huang, Y.; Zhou, J.; Duguay, F.; Chretien, N.; Welsh, E.A.; Soares, J.W.; Karatzas, C.N. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science 2002, 295, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Heidebrecht, A.; Eisoldt, L.; Diehl, J.; Schmidt, A.; Geffers, M.; Lang, G.; Scheibel, T. Biomimetic fibers made of recombinant spidroins with the same toughness as natural spider silk. Adv. Mater. 2015, 27, 2189–2194. [Google Scholar] [CrossRef] [PubMed]
- Chaw, R.C.; Correa-Garhwal, S.M.; Clarke, T.H.; Ayoub, N.A.; Hayashi, C.Y. Proteomic evidence for components of spider silk synthesis from black widow silk glands and fibers. J. Proteome Res. 2015, 14, 4223–4231. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.; Chuang, T.; Lin, A.; Joo, H.; Tsai, J.; Crawford, T.; Zhao, L.; Hsia, Y.; Williams, C.; Vierra, C.A. Dragline silk: A fiber assembled with low-molecular-weight cysteine-rich proteins. Biomacromolecules 2014, 15, 4073–4081. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.K.; Hayashi, C.Y.; Whitworth, G.B.; Ayoub, N.A. Complex gene expression in the dragline silk producing glands of the western black widow (latrodectus hesperus). BMC Genom. 2013, 14, 846. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.H.; Garb, J.E.; Hayashi, C.Y.; Haney, R.A.; Lancaster, A.K.; Corbett, S.; Ayoub, N.A. Multi-tissue transcriptomics of the black widow spider reveals expansions, co-options, and functional processes of the silk gland gene toolkit. BMC Genom. 2014, 15, 365–382. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos-Pinto, J.R.A.; Lamprecht, G.; Chen, W.; Heo, S.; Hardy, J.G.; Priewalder, H.; Scheibel, T.R.; Palma, M.S.; Lubec, G. Structure and post-translational modifications of the web silk protein spidroin-1 from Nephila spiders. J. Proteom. 2014, 105, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos-Pinto, J.R.A.; Caviquioli Garcia, A.M.; Arcuri, H.A.; Esteves, F.G.; Salles, H.C.; Lubec, G.; Palma, M.S. Silkomics: Insight into the silk spinning process of spiders. J. Proteome Res. 2016, 15, 1179–1193. [Google Scholar] [CrossRef] [PubMed]
- Gatesy, J.; Hayashi, C.; Motriuk, D.; Woods, J.; Lewis, R. Extreme diversity, conservation, and convergence of spider silk fibroin sequences. Science 2001, 291, 2603–2605. [Google Scholar] [CrossRef] [PubMed]
- Guerette, P.A.; Ginzinger, D.G.; Weber, B.H.; Gosline, J.M. Silk properties determined by gland-specific expression of a spider fibroin gene family. Science 1996, 272, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Dicko, C.; Vollrath, F.; Kenney, J.M. Spider silk protein refolding is controlled by changing pH. Biomacromolecules 2004, 5, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F. Strength and structure of spiders’ silks. J. Biotechnol. 2000, 74, 67–83. [Google Scholar] [CrossRef]
- Jin, H.J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Larracas, C. Qualitative Observation of Solubilization of Silk Fibers in Different Solvents; University of the Pacific: Stockton, CA, USA, 2016. [Google Scholar]
- Bendtsen, J.D.; Nielsen, H.; von Heijne, G.; Brunak, S. Improved prediction of signal peptides: Signalp 3.0. J. Mol. Biol. 2004, 340, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.P.; Knight, M.M.; Vollrath, F. β transition and stress-induced phase separation in the spinning of spider dragline silk. Int. J. Biol. Macromol. 2000, 27, 205–210. [Google Scholar] [CrossRef]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Madsen, B.; Shao, Z. The effect of spinning conditions on the mechanics of a spider’s dragline silk. Proc. R. Soc. Lond. B. Biol. Sci. 2001, 268, 2339–2346. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Eisoldt, L.; Hardy, J.G.; Vendrely, C.; Coles, M.; Scheibel, T.; Kessler, H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 2010, 465, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Eisoldt, L.; Thamm, C.; Scheibel, T. Review the role of terminal domains during storage and assembly of spider silk proteins. Biopolymers 2012, 97, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Young, R.J.; Vollrath, F. The effect of solvents on spider silk studied by mechanical testing and single-fibre raman spectroscopy. Int. J. Biol. Macromol. 1999, 24, 295–300. [Google Scholar] [CrossRef]
- Shao, Z.Z.; Vollrath, F. The effect of solvents on the contraction and mechanical properties of spider silk. Polymer 1999, 40, 1799–1806. [Google Scholar] [CrossRef]
- Madsen, B.; Shao, Z.Z.; Vollrath, F. Variability in the mechanical properties of spider silks on three levels: Interspecific, intraspecific and intraindividual. Int. J. Biol. Macromol. 1999, 24, 301–306. [Google Scholar] [CrossRef]
- Reed, E.J.; Bianchini, L.L.; Viney, C. Sample selection, preparation methods, and the apparent tensile properties of silkworm (B. mori) cocoon silk. Biopolymers 2012, 97, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Keten, S.; Xu, Z.; Ihle, B.; Buehler, M.J. Nanoconfinement controls stiffness, strength and mechanical toughness of β-sheet crystals in silk. Nat. Mater. 2010, 9, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Garb, J.E.; Hayashi, C.Y. Modular evolution of egg case silk genes across orb-weaving spider superfamilies. Proc. Natl. Acad. Sci. USA 2005, 102, 11379–11384. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Kohler, K.; Falick, A.M.; Moore, A.M.; Jones, P.R.; Sparkman, O.D.; Vierra, C. Egg case protein-1. A new class of silk proteins with fibroin-like properties from the spider Latrodectus hesperus. J. Biol. Chem. 2005, 280, 21220–21230. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Kohler, K.; Falick, A.M.; Moore, A.M.; Jones, P.R.; Vierra, C. Spider egg case core fibers: Trimeric complexes assembled from TuSp1, ECP-1, and ECP-2. Biochemistry 2006, 45, 3506–3516. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, F.; La Mattina, C.; Tuton-Blasingame, T.; Hsia, Y.; Gnesa, E.; Zhao, L.; Franz, A.; Vierra, C. Microdissection of black widow spider silk-producing glands. J. Vis. Exp. 2011. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larracas, C.; Hekman, R.; Dyrness, S.; Arata, A.; Williams, C.; Crawford, T.; Vierra, C.A. Comprehensive Proteomic Analysis of Spider Dragline Silk from Black Widows: A Recipe to Build Synthetic Silk Fibers. Int. J. Mol. Sci. 2016, 17, 1537. https://doi.org/10.3390/ijms17091537
Larracas C, Hekman R, Dyrness S, Arata A, Williams C, Crawford T, Vierra CA. Comprehensive Proteomic Analysis of Spider Dragline Silk from Black Widows: A Recipe to Build Synthetic Silk Fibers. International Journal of Molecular Sciences. 2016; 17(9):1537. https://doi.org/10.3390/ijms17091537
Chicago/Turabian StyleLarracas, Camille, Ryan Hekman, Simmone Dyrness, Alisa Arata, Caroline Williams, Taylor Crawford, and Craig A. Vierra. 2016. "Comprehensive Proteomic Analysis of Spider Dragline Silk from Black Widows: A Recipe to Build Synthetic Silk Fibers" International Journal of Molecular Sciences 17, no. 9: 1537. https://doi.org/10.3390/ijms17091537