1. Introduction
In the process of biological evolution, polyploidy (the duplication of the whole genome) is a universal and natural phenomenon. It leads to an increase of gene dosage and genome, which provides the space and potential for biological evolution. Polyploids generally exist among many plants and animals. Especially in freshwater fish, more than 30 kinds of polyploidy types have been found in China [
1,
2]. Researches in recent years show that the process of polyploidy and stabilization can lead to comprehensive changes in the structure, expression and function of genes. It affects the processes and the mechanisms of all genetic and epigenetic products [
3]. Epigenetics is the genetic change in gene expression that does not involve the occurrence of DNA sequence change, which has important effects on the formation and evolution of multiples. The phenomenon and mechanism of epigenetics is researched widely and related to the polyploidy incorporation of DNA methylation, gene state, nucleolar dominance, etc. [
4]. DNA methylation as a sort of modification process and reaction that commonly occurs in cells is a major epigenetic modification of genome DNA. It is also an important means of regulating gene function [
5]. In recent years, methylation-sensitive amplified polymorphism (MSAP) technology has widely been used in animal and plant genome DNA methylation level and pattern analysis. However, there are few reports on the application in fish [
6]. Loach (
Misgurnus anguillicaudatus), besides being unique fish with high economic value, has multiple reproduction patterns and a phenomenon of ploidy variation [
7]. Research shows that natural diploid, triploid, and tetraploid loaches exist in the same area in China [
8,
9,
10]. Therefore, loach is an ideal material to explore the mechanism of homologous polyploids and allopolyploids. In the last 10 years, our laboratory has devised systematic research into the distribution, origin and formation mechanism of China’s natural polyploidy loach and has investigated the cellular and molecular patterns. China’s unique natural tetraploid loach is a homologous polyploidy in which both male and female are fertile [
11,
12,
13]. At the same time, we also used natural tetraploid and diploid loach to hybridize a new triploid to research the composition of the gamete chromosomes [
14]. To the best of our knowledge, there is no report into the epigenetic mechanism among different polyploid loach, hybrid triploid loach and parents. Therefore, in this research, we selected different polyploid loach (diploid, triploid and tetraploid) and a reciprocal cross generation of diploid and tetraploid parents as subjects. Our laboratory has established an MSAP (methylation-sensitive amplified polymorphism) reaction system to explore the changes on DNA methylation levels and patterns of different polyploidy and parent–offspring loaches, and to evaluate the regulation mechanism at gene expression changes of polyploidy in loach. This study aims to explore homologous and heterologous polyploidy mechanisms in fish.
3. Discussion
After species polyploidy, homologous polyploids and allopolyploids overcome the effects of genome doubling, some genetic and epigenetic changes will soon be produced, making it faster and better adapted to the new environment. The variation of epigenetics plays an important role in improving the diversity of polyploid gene expression, inducing the diploidization of genetics and cytology and promoting mutual coordination between the genome, and so on [
16]. As an important form of epigenetic modifications, DNA methylation plays an important role in controlling gene expression, maintaining the stability of the genome, etc. Much research shows that after genome polyploidy the changes of levels and the adjustment of DNA methylation patterns is closely related to maintaining the stability of the genome, and balancing the reconstruction of nucleoplasm in polyploidy [
17,
18,
19]. Generally speaking, DNA methylation and gene expression show a negative correlation, no matter whether in diploid or polyploid, DNA demethylation can lead to gene activation or translocation activation. High levels of methylation always lead to certain gene silencing or inhibition, expression and the expression level of related gene are likely to change [
20]. In studies of
Arabidopsis thaliana [
21,
22], when the genome DNA is at high level of methylation, it has been discovered that the expression of some genes are silenced. When methylation of DNA is processed, the level of methylation is reduced, and the related gene silencing phenomena is then lifted [
19]. As a result, the relation of ploidy and DNA methylation levels in the previous are not the same. There are three main trends: first, along with the increase of ploidy, DNA methylation levels gradually increase, which shows a positive correlation [
16,
23]; second, along with the increase of ploidy, DNA methylation levels gradually reduce, which show a negative correlation [
24,
25]; and, third, no specific rule is shown [
26,
27,
28].
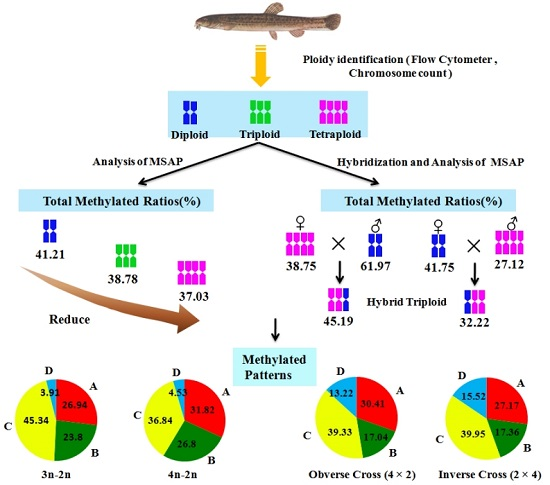
In this research, DNA methylation levels of different ploidy also change; the total methylation rates of diploid, triploid and tetraploid were 42.21%, 38.78% and 37.03%, respectively. Methylation levels show a significant difference between triploid, tetraploid and diploid loach (
p < 0.01). However, there was no significant difference between the triploid and tetraploid loach. This indicates that in the process of loach doubling its genome DNA methylation modification changes have taken place. The study found that the polyploid methylation levels did not increase with increasing polyploid levels, but gradually decreased with the increase of polyploid levels. This belongs to the second type of characteristic. After loach polyploidy, because of the increased genome, the methylation level is relatively lower, which means that some gene silencing is lifted, then related transcription can be reactivated. This means that the polyploid loach can regulate the redundant genes by DNA methylation. It also indicates that auto tetraploid loach has greater improvement potential and wider adaptability, shows advantageous characteristics of quick growth, low oxygen consumption rate, high nutritional value, etc. [
29]. Many adjustments of polyploid methylation patterns can induce the activation and silencing of certain genes, leading to changes of epigenetics, making the polyploid better adapted to the needs of the environment. In this experiment, based on analyzing changes of different ploidy on loach genomic DNA methylation patterns, it was found that, compared with the diploid loach, methylation patterns of tetraploid sites have changed 68.17% (hypermethylation 36.84%, demethylation 26.80%, and hypomethylation 4.53%), and the triploid sites have changed 73.05% (hypermethylation 45.34%, demethylation 23.80%, and hypomethylation 3.91%). It can be seen that the proportion of hypermethylation gene is significantly higher than the proportion of demethylation. Polyploid loach, possibly through methylation of some functional genes, does not express to mitigate the effects of genome double dose effect. In research with wheat [
30], rice [
26], and
Stevia rebaudiana [
25], conclusions are basically identical.
Triploid fish have characteristics of infertility, fast growth, good meat quality, disease resistance etc.; they also have great significance in breeding [
31]. An increasing number of studies show that the regulation of genetic level is conducive to the stability of hybridization and the genome doubling polyploid. Furthermore, the epigenetic modification, in which DNA methylation is the main mechanism, is very important for species formation and the successful evolution of multiple hybrid organisms. With the deepening research of DNA methylation, there are some reports about the relationship between heterosis and DNA methylation in plants. Romagnoli et al. [
32] first put forward the relationship between heterosis and gene expression in crops, and found that 33% of specific expression products re-express. The study of triploid loquat and its parent by Wang [
33] shows that the change of DNA methylation can promote the formation of triploid loquat heterosis. The study of hybrid larch species by Li et al. [
34] shows that the formation of heterosis is related to the significant increase of DNA methylation in the offspring. The study of dandelion by Verhoeven et al. [
35] compared MSAP and amplified fragment length polymorphism (AFLP) fragment inheritance in a diploid and triploid cross, and revealed de novo methylation variation between triploid F
1 individuals. However, research in aquatic animals is relatively deficient. The research of DNA methylation patterns in red crucian carp and allotetraploid crucian carp through the MSAP method used by Song et al. [
36] showed that allotetraploid crucian carp inherit 61.69% of their methylation pattern from both parents or one of them, indicating that the methylation level mainly follows the laws of Mendelian inheritance and maintains stable methylation patterns in tetraploid generations. The hybrid progeny of Zhi Kong scallops and Japanese scallops have 19.98% of sites in the methylation state, as reported by Yu et al. [
37]. Cao et al. [
6], through MSAP analysis of 20 full-sib in grass carp, found that methylation sites accounted for 75.9% of total sites. For triploid loach, whether it is a naturally occurring or synthetic, in the process of triploid formation from different individual genome reorganization and double phenomenon, to regain the balance of the gene of unpaired chromosomes, it cannot through large-scale chromosomal evolution, restore the chromosome diploidy in a short period of time. Therefore, it may regulate the extra genes and turning on and off gene expression through the changes of DNA methylation level and pattern. This study of methylation level and pattern analysis of diploid and tetraploid loach and reciprocal cross progeny shows that DNA methylation level and pattern of reciprocal cross F1 generation have obvious changes to parents. The DNA methylation level of hybrid F1 generation is between its parents, less than male parent diploid, and higher than female parent tetraploid. The hemimethylation and total methylation of reciprocal cross F1 generation is significantly higher than the orthogonal generation (
p < 0.01). In individual detection, we found that all methylation rates are greater than the hemimethylation rates in most genomes of loach. Again, the loach methylation mode is given priority to full methylation. The result that all methylation rate is higher than hemimethylation rate in mammalian genomes are the same findings of Tang et al. [
38]. The methylation pattern of reciprocal cross F1 generation have four types, namely monomorphism, demethylation, hypermethylation and hypomethylation, but they are mainly composed of type hypermethylation. This conclusion is basically identical to the research conclusion of Wang [
33], but is not the same finding in corn,
Arabidopsis thaliana [
12,
39], larch [
34] and rice [
40]. Related studies [
34] have shown that methylation means that genes translate from activation to suppression; however, demethylation means that genes translate from suppression to activation. Whether methylation or demethylation is good for heterosis, different sites of enhanced or reduced methylation have a different effect on heterosis.
4. Materials and Methods
4.1. Ethics Statement
This study was performed according to the Guide for the Care and Use of Laboratory Animals in Dalian Ocean University, Dalian, China. All animal experiments comply with Chinese laws, regulations and ethics.
4.2. Materials
Thirty diploid loaches from farmers market were used as samples in Dalian, Liaoning Province, China. All natural triploid and tetraploid loaches were from Honghu, Hubei Province, China. Sixty individuals include 30 triploid loaches and 30 tetraploid loaches. All of the loaches were fed in aquaria (22 ± 1 °C) in the laboratory of Dalian Ocean University.
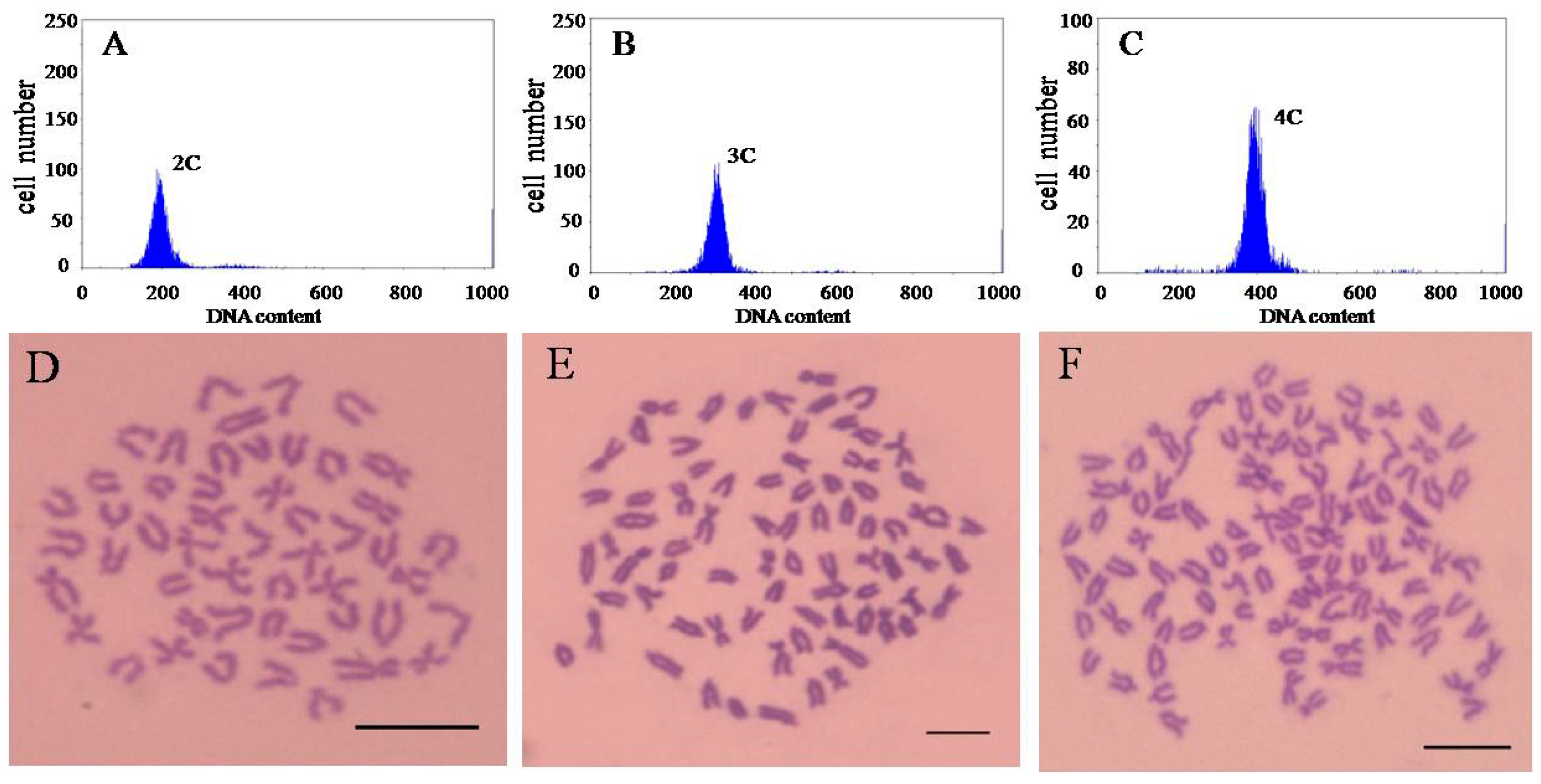
4.3. Ploidy Identification
In order to ensure the ploidy of loaches used in this experiment. The flow cytometry (Partec PAS-III, PARTEC, Münster, Germany) and chromosome number counts are used to observe ploidy detection. The specific method is as followed.
4.3.1. Flow Cytometer
Blood was collected from the caudal peduncle of loaches, and stained by DAPI (4′,6-diamidino-2-phenylindole). Using the blood DNA content of diploid loaches as a normal diploid standard, each blood sample was measured separately.
4.3.2. Preparation of Chromosome Samples
Loach was intraperitoneally injected with phytohemagglutinin (PHA; 6 μg·g−1 body weight), followed by a second PHA treatment 18–20 h later. At 4–6 h after the second PHA treatment, 0.1% colchicine (6 μg·g−1 body weight) was administered intraperitoneally. At 2–3 h after colchicines treatment, the animals were euthanized, and branchia tissue samples were collected. The branchia samples were treated under hypotonic stress in 0.8% sodium citrate for 40–45 min. The samples were fixed for 45 min in Carnoy’s solution (methanol: acetic acid = 3:1). The chromosome samples were prepared by the cold drop method and dyed by Giemsa. Statistical chromosome numbers were observed using optical microscopy.
4.4. Artificial Induce Spawning and Insemination
The parents were chosen from good development of gonad diploid and natural tetraploid loaches, injected with human chorionic gonadotropin (HCG) (injection dose: female, 20 to 25 IU·g−1; male, 10 to 12.5 IU·g−1). After 12 h, gently press female tetraploid loach abdomen, the eggs were discharged and collected in a 9 cm culture dish. Extrusion to genital pore on both sides along the male body made semen discharge. Semen was collected in centrifuge tubes by capillary (diluted 100 times with fresh water physiological saline). Using dry fertilization, hybridized combinations were obverse cross (4 × 2) and inverse cross (2 × 4). During the incubation and breeding period, the temperature was maintained at 25 ± 1 °C. Water was aerated, and dead fry were removed timely from the nursery pond. Fresh air and circulation was maintained in the breeding room.
4.5. DNA Extraction
Genomic DNA from loach fin tissue (there were 20 diploid, triploid and tetraploid loaches; 20 female and male loach of hybrid F1 generation; and four parents from obverse and inverse cross generations) was extracted by the SS-Phenol extraction method. The method was as follows. Around 0.2 g tail fin tissue was put it into 400 μL urea buffer (0.1 mol·L−1 Tris-HCl, pH: 7.5) with 10 μL proteinase K in buffer. Fin was digested for 12 h at 37 °C in a thermostat water bath. Digested fin was extracted once by saturated phenol, twice by phenol, chloroform, isoamyl alcohol mixture (volume ratio: 25:24:1), and once by chloroform. DNA was precipitated 30 min by 100% ethanol and centrifuged 10 min at speed of 12,000 rpm. The precipitation was dissolved in TE buffer (pH: 8.0) of 100 μL. The quality and concentration of DNA was tested by 1% agarose gel electrophoresis and nucleic acid concentration meter (Eppendorf Bio-Photometer D30, Eppendorf AG, Hamburg, Germany). The concentration of the various samples was adjusted for consistency, and then samples were stored at −20 °C.
4.6. Methylation-Sensitive Amplified Polymorphism (MSAP) Analysis
MSAP analysis of loach was by the MSAP technique reaction system [
41] set up by this laboratory.
4.7. Enzyme Cleavage and Adaptor Ligation
Genomic DNA was enzyme cleaved by the endonuclease
EcoR I/
Hpa II and
EcoR I/
Msp I. A total of 20 μL enzyme cleavage reaction volume included: 800 ng of genomic DNA, 4 μL of 10× buffer Tango, 10 μL of
EcoR I, and 10 μL
Hpa II or
Msp I. The reagents were thoroughly mixed and incubated 8 h at 37 °C. The adaptor ligation reaction comprised: 17 μL of enzyme cleavage product, both 1.5 μL of
EcoR I and H-M adaptor (
Table 6), 2 μL T4 DNA ligase (Transgen Biotech, Beijing China), 6 μL of 5× T4 buffer, and double-distilled water to 30 μL. Thoroughly mix reagent and incubate ligation for 2 h at 22 °C.
4.8. Preselective Amplification
The pre-amplification reaction consisted of: 4 μL adaptor ligation product, both 0.8 μL of primers E-A (10 mmol·L
−1) and H-M (10 mmol·L
−1) (
Table 6), and 1.6 μL of each dNTP (2.5 mmol·L
−1), 2 μL 10× PCR buffer (Mg
2+ free), 1.2 μL MgCl
2 (2.5 mmol·L
−1), 0.2 μL
Taq DNA polymerase (5 U·μL
−1), and double-distilled water to 20 μL. PCR conditions were as follows: initial denaturation for 2 min at 94 °C, followed by 30 cycles of denaturation for 30 s at 94 °C, annealing for 40 s at 56 °C and extension for 60 s at 72 °C, and final extension for 60 s at 72 °C. Product was temporary stored at 4 °C. The quality and concentration of pre-amplification product was tested by 1% agarose gel electrophoresis. Pre-amplification product was diluted 20 times and used as selective amplification template. Residual product was stored at −20 °C. Adaptor and primer combinations are shown in
Table 6.
4.9. Selective Amplification
Pre-amplification product was diluted 20 times in double-distilled water and was used as the template for selective amplification. The selective amplification reaction consisted of: 2 μL diluted pre-amplification product, both 1.5 μL of E (10 mmol·L
−1) and H (10 mmol·L
−1) (
Table 6), 1.5 μL of each dNTP (2.5 mmol·L
−1), 3 μL of 10× PCR buffer (Mg
2+ free), 1.2 μL of MgCl2 (2.5 mmol·L
−1), 0.2 μL
Taq DNA polymerase (5 U·μL
−1), and double-distilled water to a final volume of 20 μL. PCR conditions were as follows: initial denaturation for 2 min at 94 °C, followed by 12 cycles of denaturation for 30 s at 94 °C, annealing for 40 s at temperature from 65 to 56 °C (each cycle reduces in 0.7 °C increments) and extension for 60 s at 72 °C, followed by 30 cycles of denaturation for 40 s at 94 °C, annealing for 40 s at 56 °C and extension for 60 s at 72 °C, and final extension for 60 s at 72 °C. Product was temporary stored at 4 °C. Formamide loading buffer (10 μL) was added to the selective amplification product and the mixture was denatured for 5 min at 94 °C followed by incubation in on ice to denature the product immediately. The denatured product was tested by polyacrylamide gel electrophoresis (PAGE) and silver nitrate dying gel.
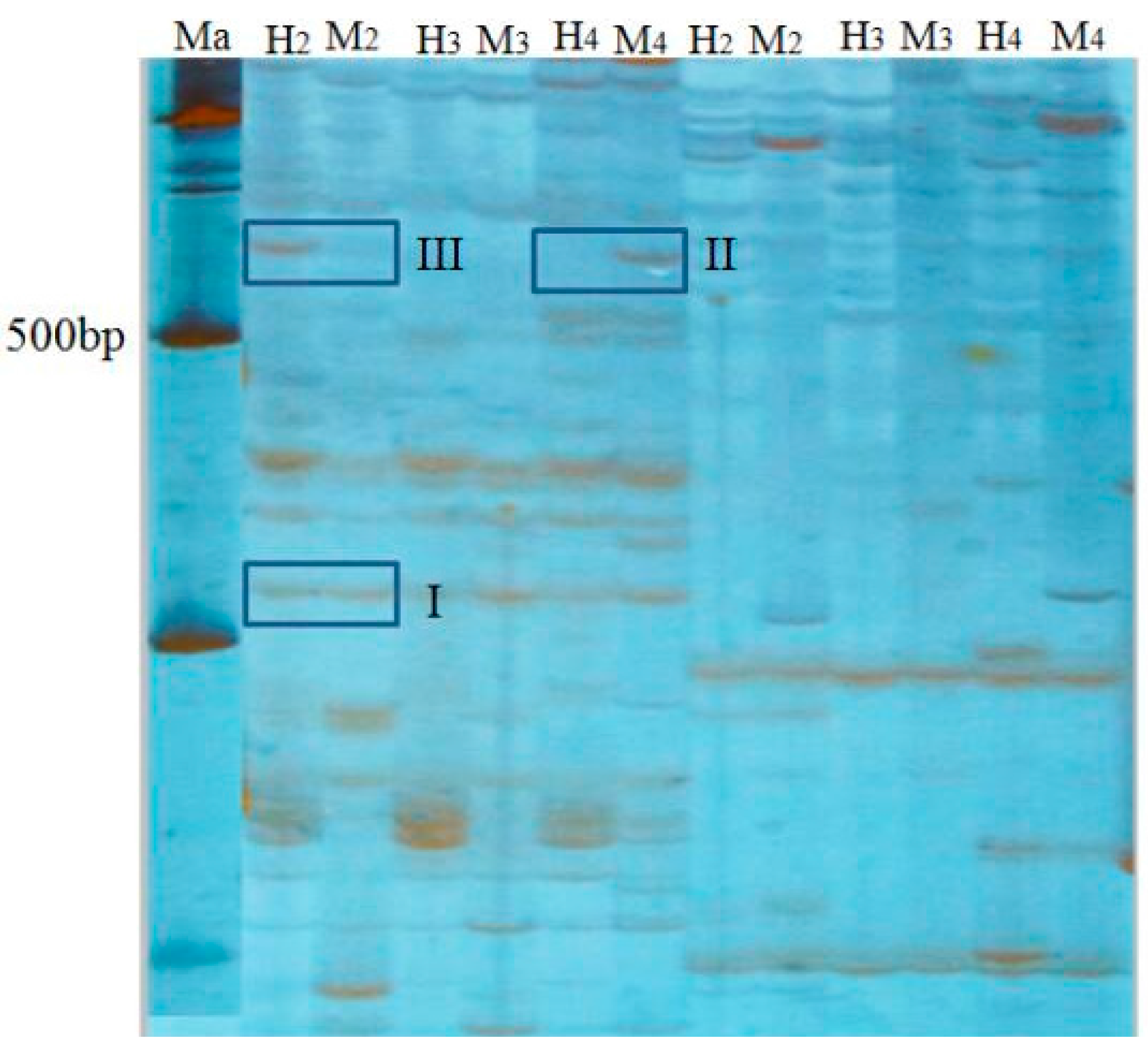
4.10. MSAP Bands Statistics, Analysis Method
Genomic DNA of parents and offspring of different ploidy loach were cleaved by two groups of endonuclease,
EcoR I/
Hpa II denotes “H”,
EcoR I/
Msp I denotes “M”. This study chose eight pairs of selective amplification primers, and counted DNA bands on an electropherogram. At the same fragment size between different lanes of one polyacrylamide gel, a visible band was noted as “1”, no band was noted as “0”. Results counted the number of bands of different banding patterns between 100 to 700 bp of product that was amplified by eight pairs of selective primers. According to whether the amplification product bands appeared in the track, methylation band type was classified into four types (
Table 7,
Figure 2). Type I, both H and M tracks have bands, indicates that the site is non-methylation. Type II, M track has band and H track is without, indicates that the site is methylated inside the DNA double-strand, also known as full methylation. Type III, H track has band and M track is without, indicates that the site is methylated outside the DNA single strand, also known as hemimethylation. Type IV, neither H nor M track have bands, indicates three cases: that site methylation is outside the DNA double-strand, or is inside and outside the DNA double-strand, or the site has no CCGG sequence. The formulas of methylation ratios were as follows: full methylation ratios (%) = full methylation bands/total bands × 100%; hemimethylation ratios (%) = hemimethylation bands/total bands × 100%; total methylation ratios = full methylation ratios + hemimethylation ratios.
4.11. Test of Significance
The test used was the Duncan’s multiple range test by Statistical Product and Service Solutions (SPSS) 19.0 (IBM, Chicago, IL, USA). p < 0.05 means significant differences and p < 0.01 means extremely significant difference.