Silk Spinning in Silkworms and Spiders
Abstract
:1. Introduction
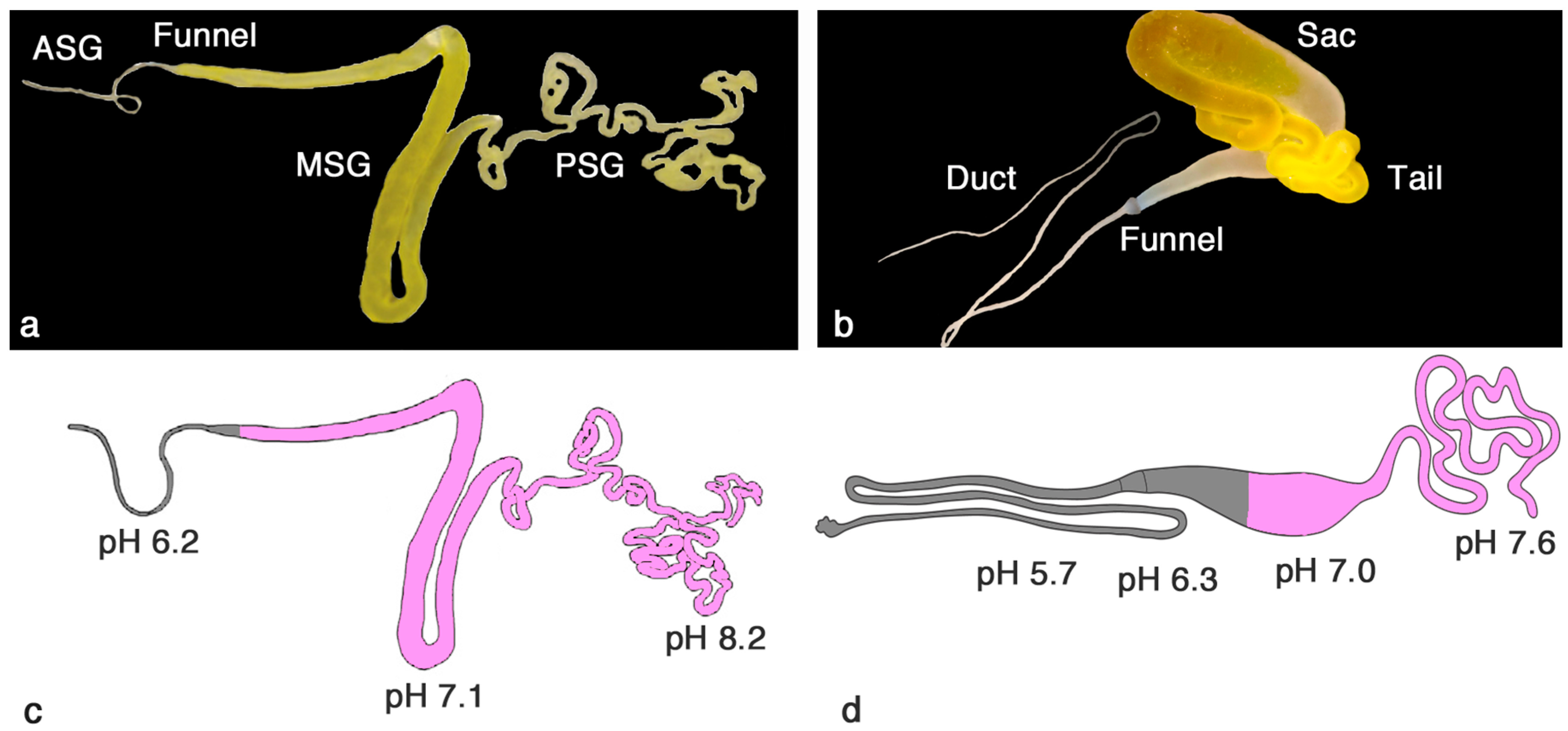
2. Similar Gland Morphologies in Silkworms and Spiders
3. pH Gradients, Ions and Shear Forces Govern Silk Formation
4. A Comparison of Fibroins and Spidroins
5. Are There Post-Translational Modifications of Silk Proteins?
6. Protein Structure Is Different in Solution and in Fibers
7. Fiber Architecture
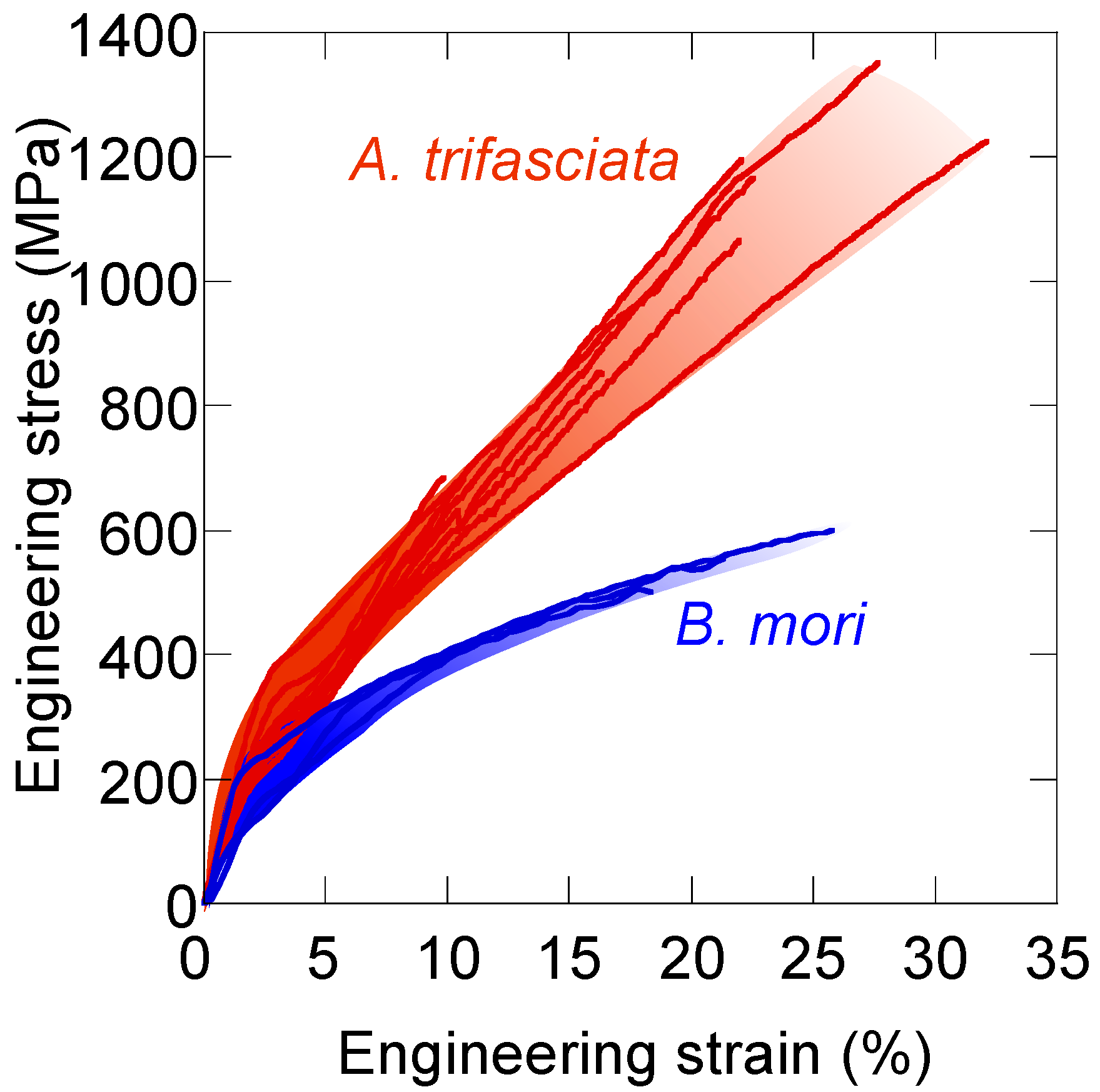
8. Tensile Properties of Silk Fibers—Why Is Spider Silk Tougher?
9. Conclusions and Future Perspectives
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PSG | Posterior silk gland |
| MSG | Middle silk gland |
| ASG | Anterior silk gland |
| CA | Carbonic anhydrase |
| Cryo-SEM-EDAX | Cryo scanning electron microscope energy dispersive X-ray |
| NT | N-terminal domain |
| CT | C-terminal domain |
| FibNT | Heavy chain fibroin N-terminal domain |
| FibCT | Heavy chain fibroin C-terminal domain |
| MaSp | Major ampullate spidroin |
| SpNT | Spidroin N-terminal domain |
| SpCT | Spidroin C-terminal domain |
| SpRep | Spidroin repetitive region |
| FibRep | Heavy chain fibroin repetitive region |
| PTM | Post-translational modification |
| NMR | Nuclear magnetic resonance |
References
- Omenetto, F.G.; Kaplan, D.L. New opportunities for an ancient material. Science 2010, 329, 528–531. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Barth, P.; Basedow, A.; Engstrom, W.; List, H. Local tolerance to spider silks and protein polymers in vivo. In Vivo 2002, 16, 229–234. [Google Scholar] [PubMed]
- Radtke, C.; Allmeling, C.; Waldmann, K.H.; Reimers, K.; Thies, K.; Schenk, H.C.; Hillmer, A.; Guggenheim, M.; Brandes, G.; Vogt, P.M. Spider silk constructs enhance axonal regeneration and remyelination in long nerve defects in sheep. PLoS ONE 2011, 6, e16990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allmeling, C.; Jokuszies, A.; Reimers, K.; Kall, S.; Choi, C.Y.; Brandes, G.; Kasper, C.; Scheper, T.; Guggenheim, M.; Vogt, P.M. Spider silk fibres in artificial nerve constructs promote peripheral nerve regeneration. Cell Prolif. 2008, 41, 408–420. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Young, J.H.; Weisman, S.; Hayashi, C.Y.; Merritt, D.J. Insect silk: One name, many materials. Annu. Rev. Entomol. 2010, 55, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Sanggaard, K.W.; Bechsgaard, J.S.; Fang, X.; Duan, J.; Dyrlund, T.F.; Gupta, V.; Jiang, X.; Cheng, L.; Fan, D.; Feng, Y.; et al. Spider genomes provide insight into composition and evolution of venom and silk. Nat. Commun. 2014, 5, 3765. [Google Scholar] [PubMed]
- Asakura, T.; Umemura, K.; Nakazawa, Y.; Hirose, H.; Higham, J.; Knight, D. Some observations on the structure and function of the spinning apparatus in the silkworm Bombyx mori. Biomacromolecules 2007, 8, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.J.; Andersson, M.; Alberti, K.A.; Chesler, M.; Xu, Q.; Johansson, J.; Rising, A.; Kaplan, D.L. Carbonic anhydrase generates a pH gradient in Bombyx mori silk glands. Insect Biochem. Mol. Biol. 2015, 65, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Holm, L.; Ridderstrale, Y.; Johansson, J.; Rising, A. Morphology and composition of the spider major ampullate gland and dragline silk. Biomacromolecules 2013, 14, 2945–2952. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.; Chen, G.; Otikovs, M.; Landreh, M.; Nordling, K.; Kronqvist, N.; Westermark, P.; Jornvall, H.; Knight, S.; Ridderstrale, Y.; et al. Carbonic anhydrase generates CO2 and H+ that drive spider silk formation via opposite effects on the terminal domains. PLoS Biol. 2014, 12, e1001921. [Google Scholar] [CrossRef] [PubMed]
- Kovoor, J. Iv comparative structure and histochemistry of silk-producing organs in arachnids. In Ecophysiology of Spiders; Nentwig, W., Ed.; Springer: Berlin, Germany, 1987; pp. 160–186. [Google Scholar]
- Rising, A.; Johansson, J. Toward spinning artificial spider silk. Nat. Chem. Biol. 2015, 11, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Peakall, D.B. Synthesis of silk, mechanism and location. Am. Zool. 1969, 9, 71–79. [Google Scholar] [CrossRef]
- Gamo, T.; Inokuchi, T.; Laufer, H. Polypeptides of fibroin and sericin secreted from the different sections of the silk gland in Bombyx mori. Insect Biochem. 1977, 7, 285–295. [Google Scholar] [CrossRef]
- Hijirida, D.H.; Do, K.G.; Michal, C.; Wong, S.; Zax, D.; Jelinski, L.W. 13C NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 1996, 71, 3442–3447. [Google Scholar] [CrossRef]
- Vollrath, F.; Knight, D.P. Liquid crystalline spinning of spider silk. Nature 2001, 410, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Magoshi, J.; Magoshi, Y.; Nakamura, S. Mechanism of fiber formation of silkworm. In Silk Polymers; American Chemical Society: Saint Louis, MO, USA, 1993; pp. 292–310. [Google Scholar]
- Davies, G.J.; Knight, D.P.; Vollrath, F. Structure and function of the major ampullate spinning duct of the golden orb weaver, Nephila edulis. Tissue Cell 2013, 45, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Knight, D.P. Structure and function of the silk production pathway in the spider Nephila edulis. Int. J. Biol. Macromol. 1999, 24, 243–249. [Google Scholar] [CrossRef]
- Breslauer, D.N.; Lee, L.P.; Muller, S.J. Simulation of flow in the silk gland. Biomacromolecules 2009, 10, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.P.; Vollrath, F. Liquid crystals and flow elongation in a spider’s silk production line. Proc. R. Soc. Lond. 1999, 266, 519–523. [Google Scholar] [CrossRef]
- Foo, C.W.P.; Bini, E.; Hensman, J.; Knight, D.P.; Lewis, R.V.; Kaplan, D.L. Role of pH and charge on silk protein assembly in insects and spiders. Appl. Phys. Mater. Sci. Process. 2006, 82, 223–233. [Google Scholar] [CrossRef]
- Miyake, S.; Azuma, M. Acidification of the silk gland lumen in Bombyx mori and samia cynthia ricini and localization of a H+-translocating vacuolar-type ATPpase. J. Insect Biotechnol. Sericol. 2008, 77, 9–16. [Google Scholar]
- Knight, D.P.; Vollrath, F. Changes in element composition along the spinning duct in a nephila spider. Naturwissenschaften 2001, 88, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Dicko, C.; Vollrath, F.; Kenney, J.M. Spider silk protein refolding is controlled by changing pH. Biomacromolecules 2004, 5, 704–710. [Google Scholar] [CrossRef] [PubMed]
- Azuma, M.; Ohta, Y. Changes in H+-translocating vacuolar-type atpase in the anterior silk gland cell of Bombyx mori during metamorphosis. J. Exp. Biol. 1998, 201, 479–486. [Google Scholar] [PubMed]
- Vollrath, F.; Knight, D.P.; Hu, X.W. Silk production in a spider involves acid bath treatment. Proc. R. Soc. Lond. 1998, 265, 817–820. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, X.; Shao, Z.; Zhou, P.; Knight, D.P.; Vollrath, F. Copper in the silk formation process of Bombyx mori silkworm. FEBS Lett. 2003, 554, 337–341. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, X.; Shao, Z.; Huang, Y.; Knight, D.P. Effect of metallic ions on silk formation in the mulberry silkworm, Bombyx mori. J. Phys. Chem. B 2005, 109, 16937–16945. [Google Scholar] [CrossRef] [PubMed]
- Knight, D.P.; Knight, M.M.; Vollrath, F. Beta transition and stress-induced phase separation in the spinning of spider dragline silk. Int. J. Biol. Macromol. 2000, 27, 205–210. [Google Scholar] [CrossRef]
- Moriya, M.; Roschzttardtz, F.; Nakahara, Y.; Saito, H.; Masubuchi, Y.; Asakura, T. Rheological properties of native silk fibroins from domestic and wild silkworms, and flow analysis in each spinneret by a finite element method. Biomacromolecules 2009, 10, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; Tanaka, K.; Arisaka, F.; Kimura, S.; Ohtomo, K.; Mizuno, S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J. Biol. Chem. 2000, 275, 40517–40528. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Z.; Confalonieri, F.; Medina, N.; Zivanovic, Y.; Esnault, C.; Yang, T.; Jacquet, M.; Janin, J.; Duguet, M.; Perasso, R.; et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 2000, 28, 2413–2419. [Google Scholar] [CrossRef] [PubMed]
- He, Y.X.; Zhang, N.N.; Li, W.F.; Jia, N.; Chen, B.Y.; Zhou, K.; Zhang, J.; Chen, Y.; Zhou, C.Z. N-terminal domain of Bombyx mori fibroin mediates the assembly of silk in response to pH decrease. J. Mol. Biol. 2012, 418, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Z.; Confalonieri, F.; Jacquet, M.; Perasso, R.; Li, Z.G.; Janin, J. Silk fibroin: Structural implications of a remarkable amino acid sequence. Proteins 2001, 44, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Tanaka, K.; Kikuchi, Y.; Waga, M.; Waga, S.; Mizuno, S. Production of a chimeric fibroin light-chain polypeptide in a fibroin secretion-deficient naked pupa mutant of the silkworm Bombyx mori. J. Mol. Biol. 1995, 251, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Long, D.; Lu, W.; Zhang, Y.; Guo, Q.; Xiang, Z.; Zhao, A. New insight into the mechanism underlying fibroin secretion in silkworm, Bombyx mori. FEBS J. 2015, 282, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Q.; Priewalder, H.; John, J.P.; Lubec, G. Silk cocoon of Bombyx mori: Proteins and posttranslational modifications—Heavy phosphorylation and evidence for lysine-mediated cross links. Proteomics 2010, 10, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Rising, A.; Hjalm, G.; Engstrom, W.; Johansson, J. N-terminal nonrepetitive domain common to dragline, flagelliform, and cylindriform spider silk proteins. Biomacromolecules 2006, 7, 3120–3124. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Liu, X.; Zhang, Y.; Lin, S.; Yang, Z.; Johansson, J.; Rising, A.; Meng, Q. Full-length minor ampullate spidroin gene sequence. PLoS ONE 2012, 7, e52293. [Google Scholar] [CrossRef] [PubMed]
- Askarieh, G.; Hedhammar, M.; Nordling, K.; Saenz, A.; Casals, C.; Rising, A.; Johansson, J.; Knight, S.D. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature 2010, 465, 236–238. [Google Scholar] [CrossRef] [PubMed]
- Gaines, W.A.; Sehorn, M.G.; Marcotte, W.R., Jr. Spidroin N-terminal domain promotes a pH-dependent association of silk proteins during self-assembly. J. Biol. Chem. 2010, 285, 40745–40753. [Google Scholar] [CrossRef] [PubMed]
- Kronqvist, N.; Otikovs, M.; Chmyrov, V.; Chen, G.; Andersson, M.; Nordling, K.; Landreh, M.; Sarr, M.; Jornvall, H.; Wennmalm, S.; et al. Sequential pH-driven dimerization and stabilization of the N-terminal domain enables rapid spider silk formation. Nat. Commun. 2014, 5, 3254. [Google Scholar] [CrossRef] [PubMed]
- Otikovs, M.; Chen, G.; Nordling, K.; Landreh, M.; Meng, Q.; Jornvall, H.; Kronqvist, N.; Rising, A.; Johansson, J.; Jaudzems, K. Diversified structural basis of a conserved molecular mechanism for pH-dependent dimerization in spider silk n-terminal domains. Chembiochem 2015, 16, 1720–1724. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.; Leclerc, J.; Lefevre, T.; Gagne, S.M.; Auger, M. Effect of pH on the structure of the recombinant C-terminal domain of Nephila clavipes dragline silk protein. Biomacromolecules 2014, 15, 4447–4454. [Google Scholar] [CrossRef] [PubMed]
- Hagn, F.; Eisoldt, L.; Hardy, J.G.; Vendrely, C.; Coles, M.; Scheibel, T.; Kessler, H. A conserved spider silk domain acts as a molecular switch that controls fibre assembly. Nature 2010, 465, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Maurer-Stroh, S.; Debulpaep, M.; Kuemmerer, N.; Lopez de la Paz, M.; Martins, I.C.; Reumers, J.; Morris, K.L.; Copland, A.; Serpell, L.; Serrano, L.; et al. Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods 2010, 7, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Tjernberg, L.O.; Rising, A.; Johansson, J.; Jaudzems, K.; Westermark, P. Transmissible amyloid (Review Symposium). J. Intern. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Escamilla, A.M.; Rousseau, F.; Schymkowitz, J.; Serrano, L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol. 2004, 22, 1302–1306. [Google Scholar] [CrossRef] [PubMed]
- Linding, R.; Schymkowitz, J.; Rousseau, F.; Diella, F.; Serrano, L. A comparative study of the relationship between protein structure and β-aggregation in globular and intrinsically disordered proteins. J. Mol. Biol. 2004, 342, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, F.; Schymkowitz, J.; Serrano, L. Protein aggregation and amyloidosis: Confusion of the kinds? Curr. Opin. Struct. Biol. 2006, 16, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lewis, R.V. Structure of a protein superfiber: Spider dragline silk. Proc. Natl. Acad Sci. USA 1990, 87, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.; Dong, Z.; Xu, M.; Lewis, R.V. Spider silk: A mystery starting to unravel. Results Probl. Cell Differ. 1992, 19, 227–254. [Google Scholar] [PubMed]
- Rising, A. Controlled assembly: A prerequisite for the use of recombinant spider silk in regenerative medicine? Acta Biomater. 2014, 10, 1627–1631. [Google Scholar] [CrossRef] [PubMed]
- Eisoldt, L.; Thamm, C.; Scheibel, T. Review the role of terminal domains during storage and assembly of spider silk proteins. Biopolymers 2012, 97, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Scheibel, T. Spider silks: Recombinant synthesis, assembly, spinning, and engineering of synthetic proteins. Microb. Cell Factories 2004, 3, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi, D.; Tokareva, O.; Rim, N.G.; Wong, J.Y.; Kaplan, D.L.; Buehler, M.J. Silk-its mysteries, how it is made, and how it is used. ACS Biomater. Sci. Eng. 2015, 1, 864–876. [Google Scholar] [CrossRef] [PubMed]
- dos Santos-Pinto, J.R.; Lamprecht, G.; Chen, W.Q.; Heo, S.; Hardy, J.G.; Priewalder, H.; Scheibel, T.R.; Palma, M.S.; Lubec, G. Structure and post-translational modifications of the web silk protein spidroin-1 from nephila spiders. J. Proteom. 2014, 105, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos-Pinto, J.R.A.; Arcuri, H.A.; Lubec, G.; Palma, M.S. Structural characterization of the major ampullate silk spidroin-2 protein produced by the spider Nephila clavipes. Biochim. Biophys. Acta 2016. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Aso, Y.; Yamamoto, K.; Banno, Y.; Wang, Y.; Tsuchida, K.; Kawaguchi, Y.; Fujii, H. Proteome analysis of silk gland proteins from the silkworm, Bombyx mori. Proteomics 2006, 6, 2586–2599. [Google Scholar] [CrossRef] [PubMed]
- Michal, C.A.; Simmons, A.H.; Chew, B.G.; Zax, D.B.; Jelinski, L.W. Presence of phosphorus in Nephila clavipes dragline silk. Biophys. J. 1996, 70, 489–493. [Google Scholar] [CrossRef]
- Winkler, S.; Wilson, D.; Kaplan, D.L. Controlling β-sheet assembly in genetically engineered silk by enzymatic phosphorylation/dephosphorylation. Biochemistry 2000, 39, 12739–12746. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.J.; Kaplan, D.L. Mechanism of silk processing in insects and spiders. Nature 2003, 424, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Kenney, J.M.; Knight, D.; Wise, M.J.; Vollrath, F. Amyloidogenic nature of spider silk. Eur. J. Biochem. 2002, 269, 4159–4163. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Ashida, J.; Yamane, T.; Kameda, T.; Nakazawa, Y.; Ohgo, K.; Komatsu, K. A repeated β-turn structure in Poly(Ala-Gly) as a model for silk I of Bombyx mori silk fibroin studied with two-dimensional spin-diffusion NMR under off magic angle spinning and rotational echo double resonance. J. Mol. Biol. 2001, 306, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Yamazaki, T.; Aoki, A.; Shindo, H.; Asakura, T. NMR study of the structures of repeated sequences, GAGXGA (X = S, Y, V), in Bombyx mori liquid silk. Biomacromolecules 2014, 15, 104–112. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Yamane, T.; Nakazawa, Y.; Kameda, T.; Ando, K. Structure of Bombyx mori silk fibroin before spinning in solid state studied with wide angle X-ray scattering and (13)C cross-polarization/magic angle spinning nmr. Biopolymers 2001, 58, 521–525. [Google Scholar] [CrossRef]
- Hayashi, C.Y.; Shipley, N.H.; Lewis, R.V. Hypotheses that correlate the sequence, structure, and mechanical properties of spider silk proteins. Int. J. Biol. Macromol. 1999, 24, 271–275. [Google Scholar] [CrossRef]
- Asakura, T.; Yao, J. 13C CP/MAS NMR study on structural heterogeneity in Bombyx mori silk fiber and their generation by stretching. Protein Sci. 2002, 11, 2706–2713. [Google Scholar] [CrossRef] [PubMed]
- Asakura, T.; Yao, J.; Yamane, T.; Umemura, K.; Ulrich, A.S. Heterogeneous structure of silk fibers from Bombyx mori resolved by 13C solid-state NMR spectroscopy. J. Am. Chem. Soc. 2002, 124, 8794–8795. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Ohgo, K.; Sugino, R.; Kishore, R.; Asakura, T. Structural analysis of Bombyx mori silk fibroin peptides with formic acid treatment using high-resolution solid-state 13C NMR spectroscopy. Biomacromolecules 2004, 5, 1763–1769. [Google Scholar] [CrossRef] [PubMed]
- Rousseau, M.E.; Lefevre, T.; Beaulieu, L.; Asakura, T.; Pezolet, M. Study of protein conformation and orientation in silkworm and spider silk fibers using raman microspectroscopy. Biomacromolecules 2004, 5, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-P.; Feng, X.-Q.; Shi, H.-J. Variability in mechanical properties of Bombyx mori silk. Mater. Sci. Eng. C 2007, 27, 675–683. [Google Scholar] [CrossRef]
- Pérez-Rigueiro, J.; Elices, M.; Llorca, J.; Viney, C. Tensile properties of argiope trifasciata drag line silk obtained from the spider´s web. J. Appl. Polym. Sci. 2001, 82, 2245–2251. [Google Scholar] [CrossRef]
- Poza, P.; Pérez-Rigueiro, J.; Elices, M.; Llorca, J. Fractographic analysis of silkworm and spider silk. Eng. Fract. Mech. 2002, 69, 1035–1048. [Google Scholar] [CrossRef]
- Work, R.W. Duality in major ampullate silk fiber formation by Orb-web-spinning spiders. Trans. Am. Microsc. Soc. 1984, 103, 113–121. [Google Scholar] [CrossRef]
- Frische, S.; Maunsbach, A.B.; Vollrath, F. Elongate cavities and skin-core structure in nephila spider silk osberved by electron microscopy. J. Microsc. 1998, 189, 64–70. [Google Scholar] [CrossRef]
- Augsten, K.; Muhlig, P.; Herrmann, C. Glycoproteins and skin-core structure in Nephila clavipes spider silk observed by light and electron microscopy. Scanning 2000, 22, 12–15. [Google Scholar] [CrossRef] [PubMed]
- Vollrath, F.; Holtet, T.; Thogersen, H.C.; Frische, S. Structural organization of spider silk. Proc. R. Soc. Lond. 1996, 263, 147–151. [Google Scholar] [CrossRef]
- Sponner, A.; Vater, W.; Monajembashi, S.; Unger, E.; Grosse, F.; Weisshart, K. Composition and hierarchical organisation of a spider silk. PLoS ONE 2007, 2, e998. [Google Scholar] [CrossRef] [PubMed]
- Gould, S.A.; Tran, K.T.; Spagna, J.C.; Moore, A.M.; Shulman, J.B. Short and long range order of the morphology of silk from latrodectus hesperus (black widow) as characterized by atomic force microscopy. Int. J. Biol. Macromol. 1999, 24, 151–157. [Google Scholar] [CrossRef]
- Li, S.F.; McGhie, A.J.; Tang, S.L. New internal structure of spider dragline silk revealed by atomic force microscopy. Biophys. J. 1994, 66, 1209–1212. [Google Scholar] [CrossRef]
- Miller, L.D.; Putthanarat, S.; Eby, R.K.; Adams, W.W. Investigation of the nanofibrillar morphology in silk fibers by small angle X-ray scattering and atomic force microscopy. Int. J. Biol. Macromol. 1999, 24, 159–165. [Google Scholar] [CrossRef]
- Hakimi, O.; Knight, D.P.; Knight, M.M.; Grahn, M.F.; Vadgama, P. Ultrastructure of insect and spider cocoon silks. Biomacromolecules 2006, 7, 2901–2908. [Google Scholar] [CrossRef] [PubMed]
- Emile, O.; Le Floch, A.; Vollrath, F. Time-resolved torsional relaxation of spider draglines by an optical technique. Phys. Rev. Lett. 2007, 98, 167402. [Google Scholar] [CrossRef] [PubMed]
- Emile, O.; Le Floch, A.; Vollrath, F. Biopolymers: Shape memory in spider draglines. Nature 2006, 440, 621. [Google Scholar] [CrossRef] [PubMed]
- Cranford, S.W. Increasing silk fibre strength through heterogeneity of bundled fibrils. J. R. Soc. Interface 2013, 10, 20130148. [Google Scholar] [CrossRef] [PubMed]
- Plaza, G.R.; Corsini, P.; Perez-Rigueiro, J.; Marsano, E.; Guinea, G.V.; Elices, M. Effect of water on Bombyx mori regenerated silk fibers and its application in modifying their mechanical properties. J. Appl. Polym. Sci. 2008, 109, 1793–1801. [Google Scholar] [CrossRef] [Green Version]
- Plaza, G.R.; Perez-Rigueiro, J.; Riekel, C.; Perea, G.B.; Agullo-Rueda, F.; Burghammer, M.; Guinea, G.V.; Elices, M. Relationship between microstructure and mechanical properties in spider silk fibers: Identification of two regimes in the microstructural changes. Soft Matter 2012, 8, 6015–6026. [Google Scholar] [CrossRef]
- Guan, J.; Wang, Y.; Mortimer, B.; Holland, C.; Shao, Z.; Porter, D.; Vollrath, F. Glass transitions in native silk fibres studied by dynamic mechanical thermal analysis. Soft Matter 2016, 12, 5926–5936. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.; Guan, J.; Vollrath, F. Spider silk: Super material or thin fibre? Adv. Mater. 2013, 25, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Kwon, J.; Na, S. Mechanical behavior comparison of spider and silkworm silks using molecular dynamics at atomic scale. Phys. Chem. Chem. Phys. 2016, 18, 4814–4821. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, L.; Teng, P.K.; Riek, R.; Eisenberg, D. Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. USA 2010, 107, 3487–3492. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.; Nerelius, C.; Willander, H.; Presto, J. Conformational preferences of non-polar amino acid residues: An additional factor in amyloid formation. Biochem. Biophys. Res. Commun. 2010, 402, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J. Harnessing the self-assembling properties of proteins in spider silk and lung surfactant, in Amyloid Fibrils and Prefibrillar Aggregates. In Molecular and Biological Properties; Otzen, D.E., Ed.; Wiley-VCH Verlag: Weinheim, Germany, 2013; pp. 455–470. [Google Scholar]
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, J.; Andersson, H.; Nilsson, I.; White, S.H.; von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Rising, A.; Johansson, J.; Larson, G.; Bongcam-Rudloff, E.; Engstrom, W.; Hjalm, G. Major ampullate spidroins from euprosthenops australis: Multiplicity at protein, mRNA and gene levels. Insect Mol. Biol. 2007, 16, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Rockwood, D.N.; Preda, R.C.; Yucel, T.; Wang, X.; Lovett, M.L.; Kaplan, D.L. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 2011, 6, 1612–1631. [Google Scholar] [CrossRef] [PubMed]
- Hossain, K.S.; Ochi, A.; Ooyama, E.; Magoshi, J.; Nemoto, N. Dynamic light scattering of native silk fibroin solution extracted from different parts of the middle division of the silk gland of the Bombyx mori silkworm. Biomacromolecules 2003, 4, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Koebley, S.R.; Thorpe, D.; Pang, P.; Chrisochoides, P.; Greving, I.; Vollrath, F.; Schniepp, H.C. Silk reconstitution disrupts fibroin self-assembly. Biomacromolecules 2015, 16, 2796–2804. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.; Grip, S.; Rising, A.; Hedhammar, M.; Engstrom, W.; Hjalm, G.; Johansson, J. Macroscopic fibers self-assembled from recombinant miniature spider silk proteins. Biomacromolecules 2007, 8, 1695–1701. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Rainey, J.K.; Meng, Q.; Liu, X.Q. Recombinant minimalist spider wrapping silk proteins capable of native-like fiber formation. PLoS ONE 2012, 7, e50227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, F.; Jiang, X.; Cao, M.; Wang, S.; Zou, H.; Cao, Y.; Xian, M.; Liu, H. Microbial production of amino acid-modified spider dragline silk protein with intensively improved mechanical properties. Prep. Biochem. Biotechnol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Deng, Q.; Liu, X.Y.; Yang, D. Engineered large spider eggcase silk protein for strong artificial fibers. Adv. Mater. 2013, 25, 1216–1220. [Google Scholar] [CrossRef] [PubMed]
- Copeland, C.G.; Bell, B.E.; Christensen, C.D.; Lewis, R.V. Development of a process for the spinning of synthetic spider silk. ACS Biomater. Sci. Eng. 2015, 1, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Dolfe, L.; Winblad, B.; Johansson, J.; Presto, J. Brichos binds to a designed amyloid-forming β-protein and reduces proteasomal inhibition and aggresome formation. Biochem. J. 2016, 473, 167–178. [Google Scholar] [CrossRef] [PubMed]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersson, M.; Johansson, J.; Rising, A. Silk Spinning in Silkworms and Spiders. Int. J. Mol. Sci. 2016, 17, 1290. https://doi.org/10.3390/ijms17081290
Andersson M, Johansson J, Rising A. Silk Spinning in Silkworms and Spiders. International Journal of Molecular Sciences. 2016; 17(8):1290. https://doi.org/10.3390/ijms17081290
Chicago/Turabian StyleAndersson, Marlene, Jan Johansson, and Anna Rising. 2016. "Silk Spinning in Silkworms and Spiders" International Journal of Molecular Sciences 17, no. 8: 1290. https://doi.org/10.3390/ijms17081290





