The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations
Abstract
:1. Introduction
2. Results
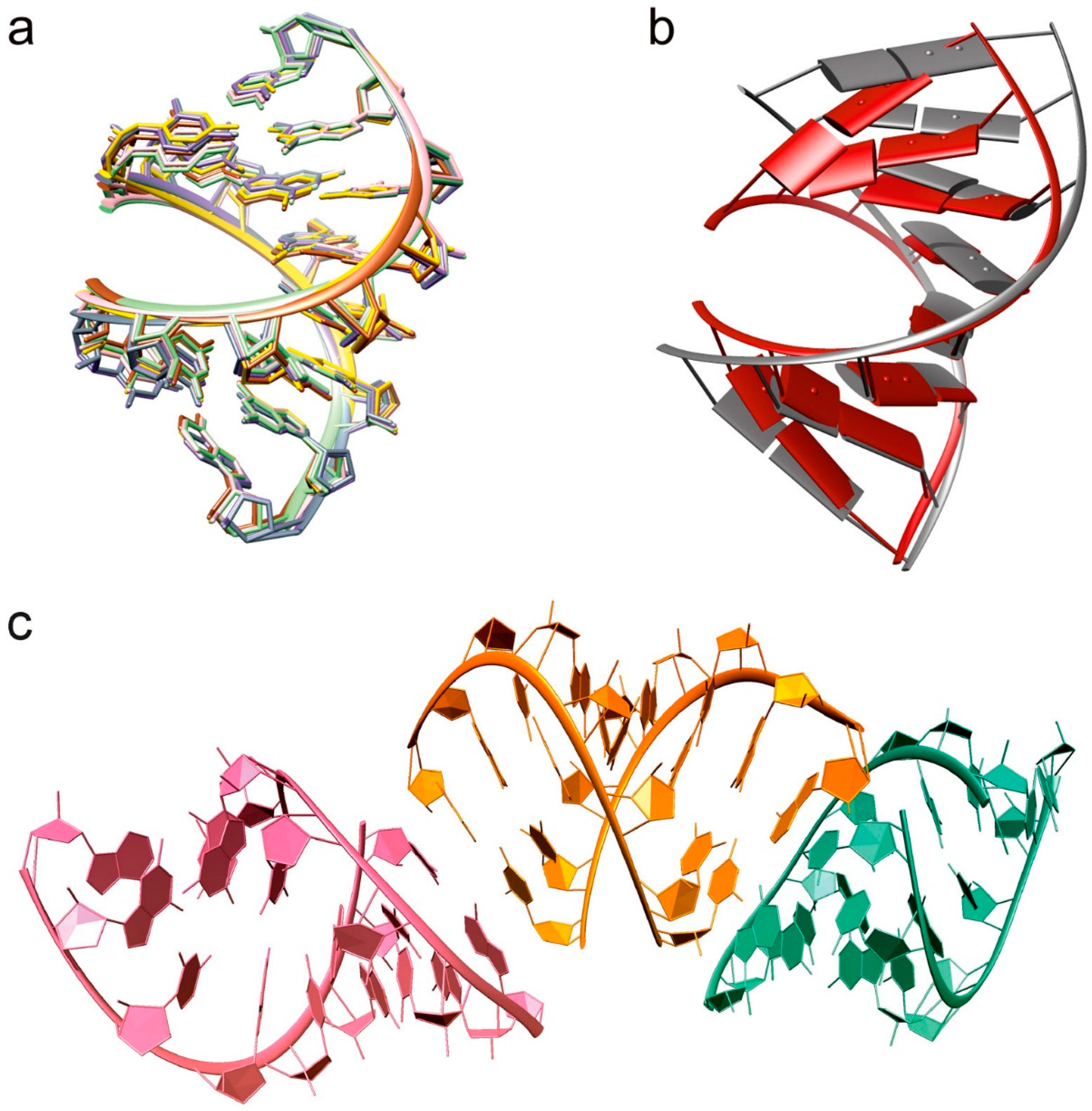
2.1. Overall Structure
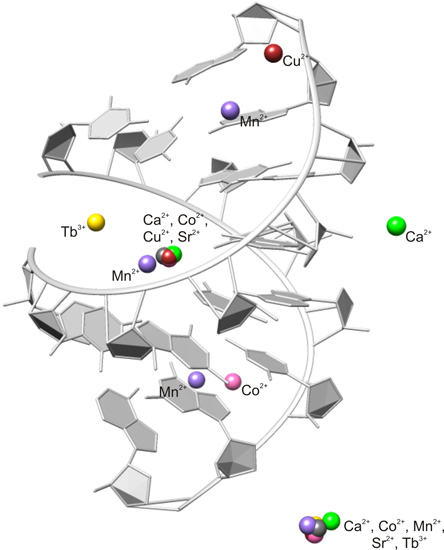
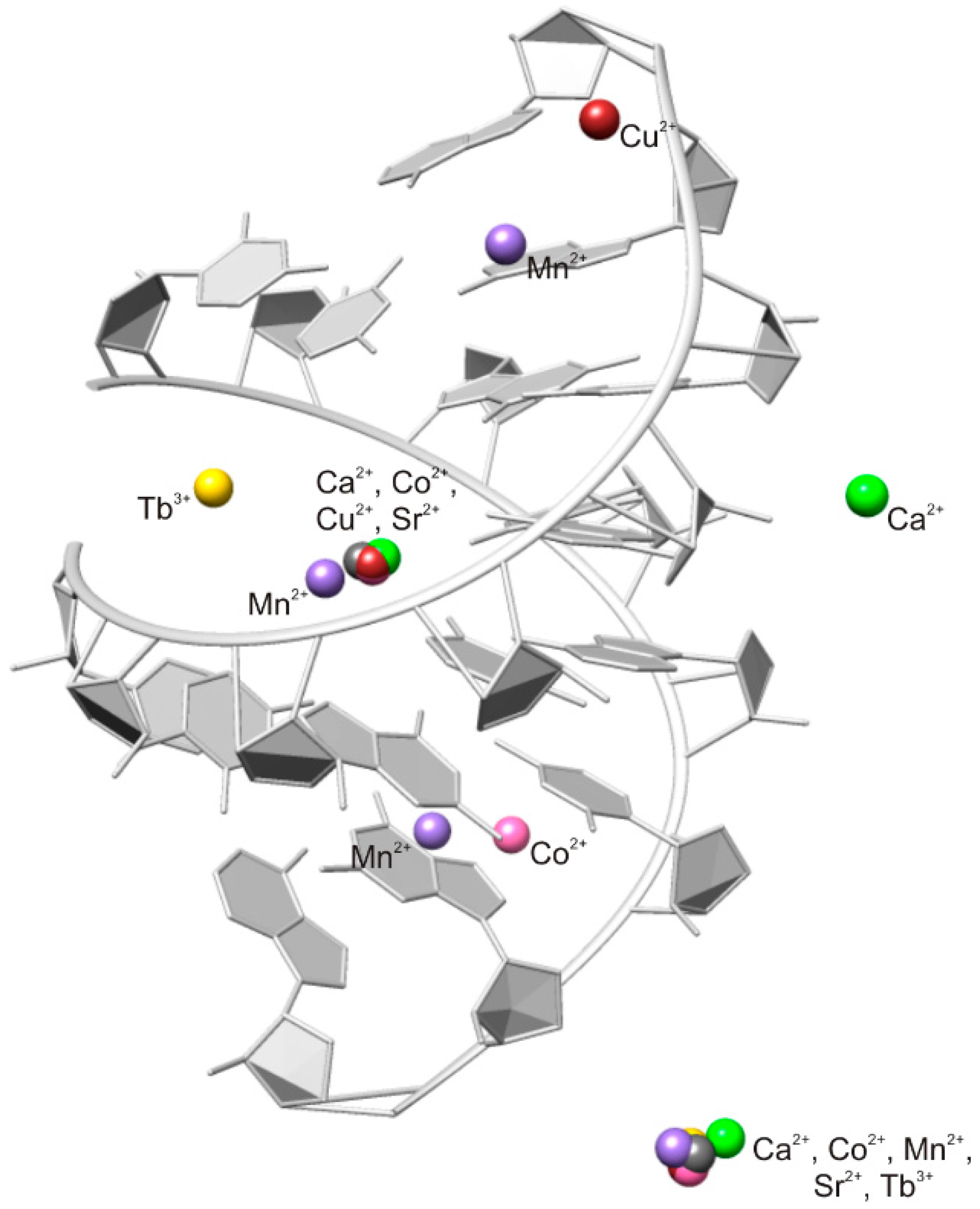
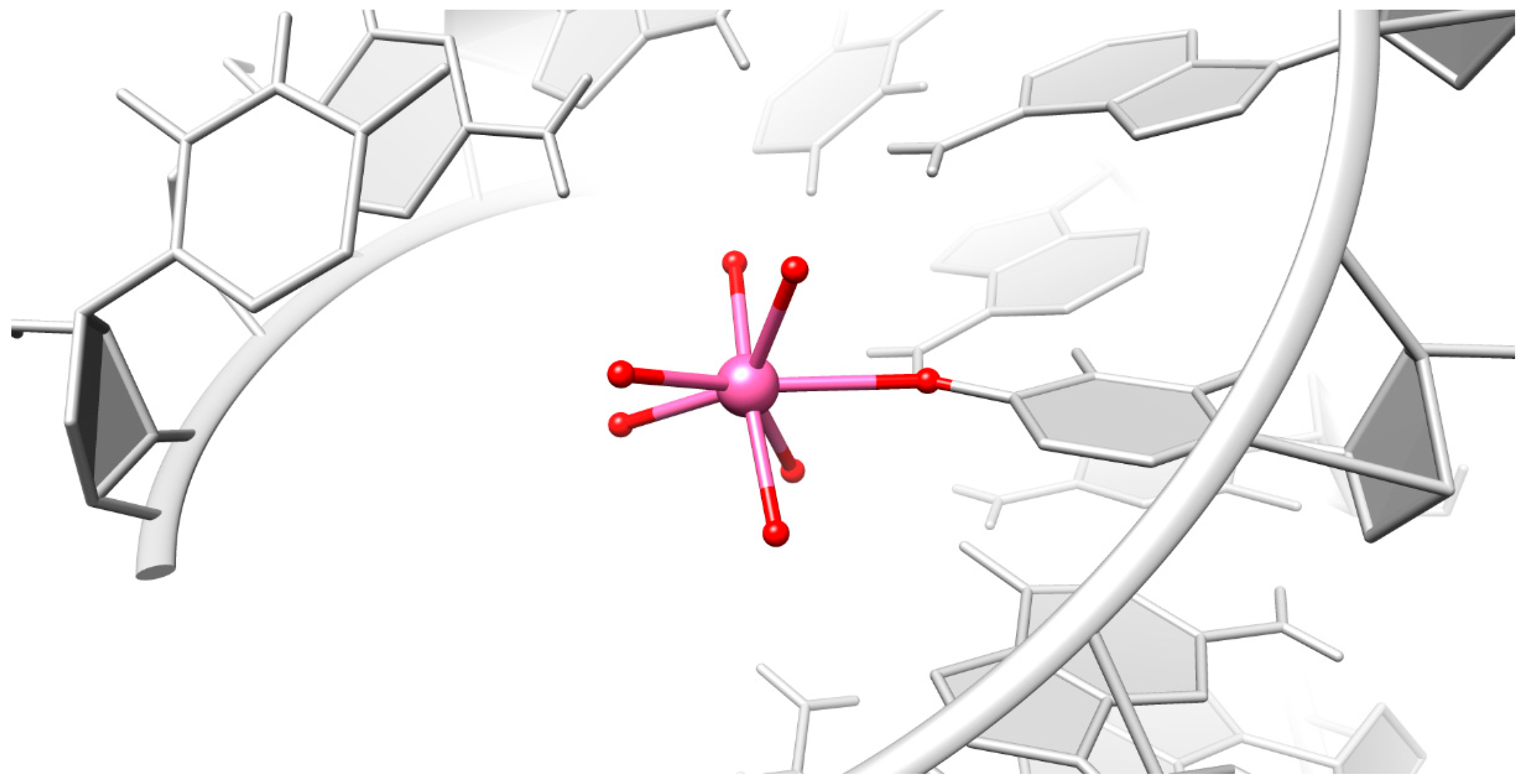
2.2. Careful Examination of the Tested Metal Ions
2.3. Comparison of Observed Metal Ion RNA Interaction to Other Macromolecules as Suggested by the MINAS Database
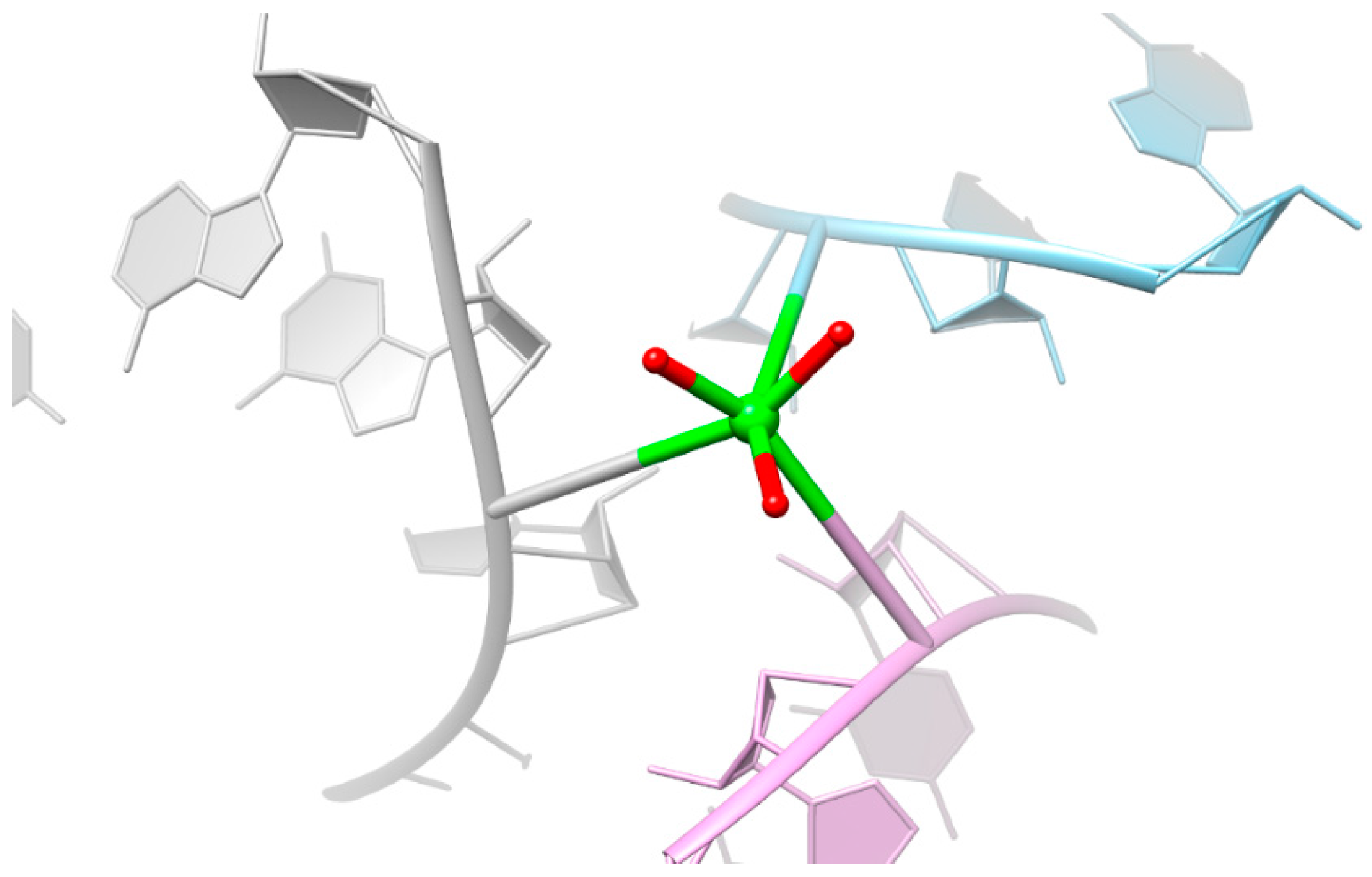
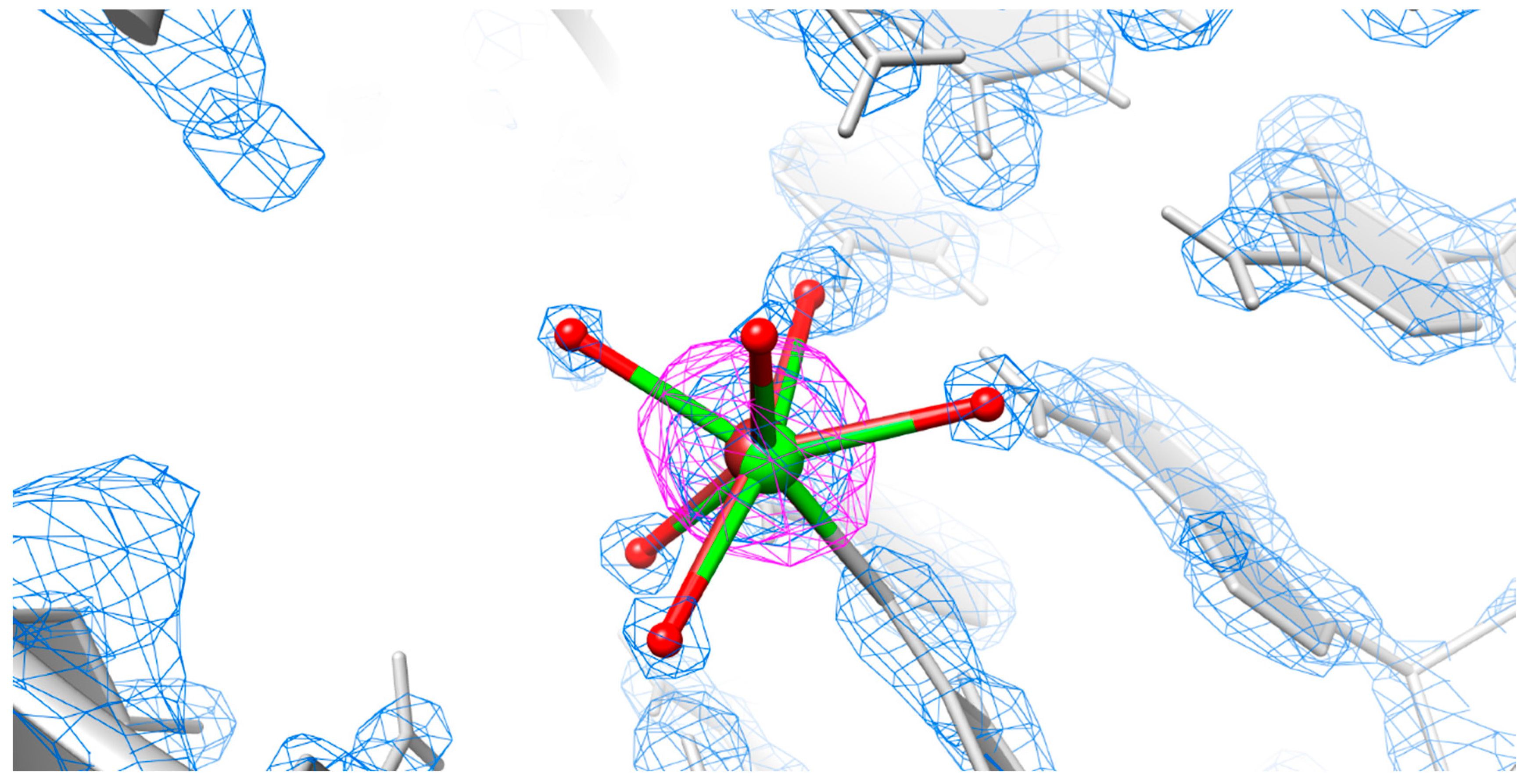
2.4. Is There Cu2+ or Ca2+ in the Copper(II) Soaked Structure?
3. Discussion
3.1. Structural Changes Are Independent of the Nature of the Cations
3.2. Predominant Localization of All Tested Metal Ions in the Phosphate Backbone and in the Central Major Groove
3.3. Particular Innersphere Binding of Co2+, Cu2+, and Ca2+ to O4 of Uracil
3.4. Mixed State Explains the Elongated Bond Length for the Copper(II) Soaked Crystal Structure
4. Methods
4.1. RNA Synthesis and Purification
4.2. RNA Crystallization and Soaking
4.3. Data Collection and Structure Determination
- List 1. Statistics of sub-structure determination and phasing (Ca2+ data).
SHELXD CCweak (%)/CCall (%) (for the top solution) 48.8/29.3 SHELXD CFOM (for the top solution) 78.1 SHELXD PATFOM (for the top solution) 18.6 Number of correct sites (for the top solution) 14 SHELXE CC (%) 69.20 SHELXE FOM 0.656 PHASER EP FOM 0.697 Map CC (%) (DM map against the map calculated from the refined model) 82.4
4.4. Localization of Metal Ions
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Protein Data Bank Accession Codes
References
- Trappl, K.; Polacek, N. The ribosome: A molecular machine powered by RNA. Met. Ions Life Sci. 2011, 9, 253–275. [Google Scholar] [PubMed]
- Freisinger, E.; Sigel, R.K.O. From nucleotides to ribozymes—A comparison of their metal ion binding properties. Coord. Chem. Rev. 2007, 251, 1834–1851. [Google Scholar] [CrossRef] [Green Version]
- Pyle, A.M. Metal ions in the structure and function of RNA. J. Biol. Inorg. Chem. 2002, 7, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Ferre-D’Amare, A.R.; Winkler, W.C. The roles of metal ions in regulation by riboswitches. Met. Ions Life Sci. 2011, 9, 141–173. [Google Scholar] [PubMed]
- Scott, W.G. RNA structure, metal ions, and catalysis. Curr. Opin. Chem. Biol. 1999, 3, 705–709. [Google Scholar] [CrossRef]
- Draper, D.E.; Grilley, D.; Soto, A.M. Ions and RNA folding. Annu. Rev. Biophys. Biomol. 2005, 34, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Lippert, B. Coordinative bond formation between metal ions and nucleic acid base. In Nucleic Acid-Metal Ion Interactions. RSC Biomolecular Science; Hud, N.V., Ed.; Royal Society of Chemistry: Cambridge, UK, 2009; pp. 39–74. [Google Scholar]
- Lipfert, J.; Doniach, S.; Das, R.; Herschlag, D. Understanding nucleic acid-ion interactions. Annu. Rev. Biochem. 2014, 83, 813–841. [Google Scholar] [CrossRef] [PubMed]
- Auffinger, P.; Grover, N.; Westhof, E. Metal ion binding to RNA. Met. Ions Life Sci. 2011, 9, 1–35. [Google Scholar] [PubMed]
- Schnabl, J.; Suter, P.; Sigel, R.K.O. MINAS—A database of Metal Ions in Nucleic Acids. Nucleic Acids Res. 2011, 40, D434–D438. [Google Scholar] [CrossRef] [PubMed]
- Erat, M.C.; Sigel, R.K.O. Divalent metal ions tune the self-splicing reaction of the yeast mitochondrial group II intron Sc.ai5γ. J. Biol. Inorg. Chem. 2008, 13, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Pechlaner, M.; Sigel, R.K.O. Characterization of Metal Ion-Nucleic Acid Interactions in Solution. Met. Ions Life Sci. 2012, 10, 1–42. [Google Scholar] [PubMed]
- Weinstein, L.B.; Jones, B.C.; Cosstick, R.; Cech, T.R. A second catalytic metal ion in group I ribozyme. Nature 1997, 388, 805–808. [Google Scholar] [PubMed]
- Gonzalez, R.L.; Tinoco, I. Identification and characterization of metal ion binding sites in RNA. Methods Enzymol. 2001, 338, 421–443. [Google Scholar] [PubMed]
- Feigon, J.; Butcher, S.E.; Finger, L.D.; Hud, N.V. Solution nuclear magnetic resonance probing of cation binding sites on nucleic acids. Methods Enzymol. 2001, 338, 400–420. [Google Scholar] [PubMed]
- Donghi, D.; Sigel, R.K.O. Metal ion–RNA interactions studied via multinuclear NMR. Methods Mol. Biol. 2012, 848, 253–273. [Google Scholar] [PubMed]
- Santangelo, M.G.; Medina-Molner, A.; Schweiger, A.; Mitrikas, G.; Spingler, B. Structural analysis of Cu(II) ligation to the 5′-GMP nucleotide by pulse EPR spectroscopy. J. Biol. Inorg. Chem. 2007, 12, 767–775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santangelo, M.G.; Antoni, P.M.; Spingler, B.; Jeschke, G. Can copper(II) mediate Hoogsteen base-pairing in a left-handed DNA duplex? A pulse EPR study. ChemPhysChem 2010, 11, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.Z.; Dieckmann, T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Chem. Biol. 2004, 14, 350–359. [Google Scholar] [CrossRef] [PubMed]
- DeRose, V.J. Metal ion binding to catalytic RNA molecules. Curr. Opin. Chem. Biol. 2003, 13, 317–324. [Google Scholar] [CrossRef]
- Duguid, J.; Bloomfield, V.A.; Benevides, J.; Thomas, G.J. Raman spectroscopy of DNA-metal complexes. I. Interactions and conformational effects of the divalent cations: Mg, Ca, Sr, Ba, Mn, Co, Ni, Cu, Pd, and Cd. Biophys. J. 1993, 65, 1916–1928. [Google Scholar] [CrossRef]
- Ascone, I.; Strange, R. Biological X-ray absorption spectroscopy and metalloproteomics. J. Synchrotron Radiat. 2009, 16, 413–421. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Chordia, M.D.; Cooper, D.R.; Chruszcz, M.; Müller, P.; Sheldrick, G.M.; Minor, W. Validation of metal-binding sites in macromolecular structures with the CheckMyMetal web server. Nat. Protoc. 2013, 9, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Ennifar, E.; Walter, P.; Dumas, P. A crystallographic study of the binding of 13 metal ions to two related RNA duplexes. Nucleic Acids Res. 2003, 31, 2671–2682. [Google Scholar] [CrossRef] [PubMed]
- Brown, I.D. Recent developments in the methods and applications of the bond valence model. Chem. Rev. 2009, 109, 6858–6919. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.A.; Brunger, A.T. The 1.8 Å crystal structure of a statically disordered 17 base-pair RNA duplex: Principles of RNA crystal packing and its effect on nucleic acid structure. J. Mol. Biol. 1999, 285, 1577–1588. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.J.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef] [PubMed]
- Popenda, M.; Szachniuk, M.; Antczak, M.; Purzycka, K.J.; Lukasiak, P.; Bartol, N.; Blazewicz, J.; Adamiak, R.W. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012, 40, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Dauter, Z.; Adamiak, D.A. Anomalous signal of phosphorus used for phasing DNA oligomer: Importance of data redundancy. Acta Crystallogr. D 2001, 57, 990–995. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Dauter, M.; Dauter, Z. Phosphates in the Z-DNA dodecamer are flexible, but their P-SAD signal is sufficient for structure solution. Acta Crystallogr. D 2014, 70, 1790–1800. [Google Scholar] [CrossRef] [PubMed]
- Sigel, R.K.O.; Sigel, H. A stability concept for metal ion coordination to single-stranded nucleic acids and affinities of individual sites. Acc. Chem. Res. 2010, 43, 974–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohner, M.; Medina-Molner, A.; Spingler, B. N,N,O, and N,O,N meridional cis coordination of two guanines to copper(II) by d(CGCGCG)2. Inorg. Chem. 2016. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Shannon, R.D.; Prewitt, C.T. Effective ionic radii in oxides and fluorides. Acta Crystallogr. B 1969, 25, 925–946. [Google Scholar] [CrossRef]
- Shannon, R.D. Revised effective ionic-radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr. A 1976, 32, 751–767. [Google Scholar] [CrossRef]
- Sigel, R.K.O. Group II intron Ribozymes and metal ions—A delicate relationship. Eur. J. Inorg. Chem. 2005, 12, 2281–2292. [Google Scholar] [CrossRef]
- Roth, A.; Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009, 78, 305–334. [Google Scholar] [CrossRef] [PubMed]
- Lehman, N.; Joyce, G.F. Evolution in vitro of an RNA enzyme with altered metal dependence. Nature 1993, 361, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Zivarts, M.; Liu, Y.; Breaker, R.R. Engineered allosteric ribozymes that respond to specific divalent metal ions. Nucleic Acids Res. 2005, 33, 622–631. [Google Scholar] [CrossRef] [PubMed]
- Sigel, H.; Griesser, R. Nucleoside 5′-triphosphates: Self-association, acid-base, and metal ion-binding properties in solution. Chem. Soc. Rev. 2005, 34, 875–900. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA 2000, 6, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Scott, W.G.; Finch, J.T.; Klug, A. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 1995, 81, 991–1002. [Google Scholar] [CrossRef]
- Scott, W.G.; Murray, J.B.; Arnold, J.R.; Stoddard, B.L.; Klug, A. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science 1996, 274, 2065–2069. [Google Scholar] [CrossRef] [PubMed]
- Marcia, M.; Pyle, A.M. Principles of ion recognition in RNA: Insights from the group II intron structures. RNA 2014, 20, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Irving, H.; Williams, R.J.P. The stability of transition-metal complexes. J. Chem. Soc. 1953, 320, 3192–3210. [Google Scholar] [CrossRef]
- Schnabl, J.; Sigel, R.K.O. Controlling ribozyme activity by metal ions. Curr. Opin. Chem. Biol. 2010, 14, 269–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigel, R.K.O.; Vaidya, A.; Pyle, A.M. Metal ion binding sites in a group II intron core. Nat. Struct. Biol. 2000, 7, 1111–1116. [Google Scholar] [PubMed]
- Cate, J.H.; Doudna, J.A. Metal-binding sites in the major groove of a large ribozyme domain. Structure 1996, 4, 1221–1229. [Google Scholar] [CrossRef]
- Knobloch, B.; Linert, W.; Sigel, H. Metal ion-binding properties of (N3)-deprotonated uridine, thymidine, and related pyrimidine nucleosides in aqueous solution. Proc. Natl. Acad. Sci. USA 2005, 102, 7459–7464. [Google Scholar] [CrossRef] [PubMed]
- Dunham, C.M.; Murray, J.B.; Scott, W.G. A helical twist-induced conformational switch activates cleavage in the hammerhead ribozyme. J. Mol. Biol. 2003, 332, 327–336. [Google Scholar] [CrossRef]
- Dahm, S.C.; Uhlenbeck, O.C. Role of divalent metal ions in the hammerhead RNA cleavage reaction. Biochemistry 1991, 30, 9464–9469. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.C.; Holbrook, A.R. Encyclopedia of Life Science, 1st ed.; Wiley: Chichester, UK, 2002. [Google Scholar]
- Gallo, S.; Furler, M.; Sigel, R.K.O. In vitro transcription and purification of RNAs of different size. Chimia 2005, 59, 812–816. [Google Scholar] [CrossRef] [Green Version]
- Kabsch, W. XDS. Acta Crystallogr. D 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Potterton, E.; Briggs, P.; Turkenburg, M.; Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D 2003, 59, 1131–1137. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.R.; Murshudov, G.N. How good are my data and what is the resolution? Acta Cristallogr. D 2013, 69, 1204–1214. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.; Wang, M.; Schulze-Briese, C. Optimal fine phi-slicing for single-photon-counting pixel detectors. Acta Crystallogr. D 2012, 68, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Shields, G.P.; Raithby, P.R.; Allen, F.H.; Motherwell, W.D.S. The assignment and validation of metal oxidation states in the Cambridge Structural Database. Acta Crystallogr. B 2000, 56, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Jones, T.A. xdlMAPMAN and xdlDATAMAN—Programs for reformatting, analysis and manipulation of biomacromolecular electron-density maps and reflection data sets. Acta Crystallogr. D 1996, 52, 826–828. [Google Scholar] [CrossRef] [PubMed]
| Calculated 8mer Duplex | Ca2+ | Mn2+ | Co2+ | Cu2+ | Sr2+ | Tb3+ | |
|---|---|---|---|---|---|---|---|
| Helical rise (Å) | 2.80 (0.03) | 2.4 (0.57) | 2.59 (0.47) | 2.41 (0.55) | 2.43 (0.54) | 2.58 (0.36) | 2.56 (0.42) |
| Helical twist (°) | 16.0 (0.9) | 36.0 (2.7) | 35.0 (2.4) | 35.6 (2.9) | 35.8 (3.1) | 35.3 (1.8) | 35.7 (1.7) |
| Major groove width (Å) | 12.7 | 5.4 | 7.9 | 5.5 | 5.6 | 7.7 | 6.3 |
| Metal ID | Cation | Inner Sphere Ligand a | Distance (Å) | Occupancy | B-Factor | Outer Sphere Ligand b | Valence c | % in MINAS |
|---|---|---|---|---|---|---|---|---|
| 1Ca/C1 | Ca2+ | 1.0 | 19.4 | 2.2 | ||||
| O4 4U/A | 2.39 | 1.0 | 18.1 | 0.7 | ||||
| O 2HOH/D | 2.43 | 1.0 | 21.7 | N7 3G/A | 3.9 | |||
| O 3HOH/D | 2.45 | 1.0 | 21.5 | |||||
| O 14HOH/D | 2.39 | 1.0 | 21.5 | O6 3G/A | 2.9 | |||
| O 15HOH/D | 2.36 | 1.0 | 23.6 | |||||
| O 16HOH/D | 2.35 | 1.0 | 18.4 | O6 3G/B | 2.9 | |||
| O 17HOH/D | 2.4 | 1.0 | 18.1 | |||||
| 1Ca/E1 | Ca2+ | 0.4 | 21.7 | |||||
| O2′/5 A/B | 2.86 | 1.0 | 22.6 | 2.1 | ||||
| O2′/5 A/B d | 2.86 | 1.0 | 22.6 | 2.1 | ||||
| O2′/5 A/B d | 2.86 | 1.0 | 22.6 | 2.1 | ||||
| 1Ca/F1 | Ca2+ | 1.0 | 18.3 | 1.8 | ||||
| OP1 7G/A | 2.34 | 1.0 | 21.0 | 18.7 | ||||
| OP1 7G/A d | 2.34 | 1.0 | 21.0 | 18.7 | ||||
| OP1 7G/A d | 2.34 | 1.0 | 21.0 | 18.7 | ||||
| O 4HOH/D | 2.52 | 1.0 | 26.1 | |||||
| O 4HOH/D d | 2.52 | 1.0 | 26.1 | |||||
| O 4HOH/D d | 2.52 | 1.0 | 26.1 | |||||
| 1Cu/C1 | Cu2+ | 0.4 0.46 e | 19.1 | |||||
| O4 4U/A | 2.40 | 1.0 | 18.1 | 0.0 (Cu2+) 0.7 (Ca2+) | ||||
| O 18HOH/D | 2.32 | 1.0 | 19.1 | O6 3G/A | 10.3 (Cu2+) 2.9 (Ca2+) | |||
| O 22HOH/D | 2.30 | 1.0 | 18.3 | O6 3G/A | 10.3 (Cu2+) 2.9 (Ca2+) | |||
| O 7HOH/D | 2.38 | 1.0 | 22.9 | |||||
| O 5HOH/D | 2.38 | 1.0 | 20.9 | N7 3G/A | 4.8 (Cu2+) 3.9 (Ca2+) | |||
| O 2HOH/D | 2.31 | 1.0 | 21.0 | N4 2C/B | 9.6 (Cu2+) 0.5 (Ca2+) | |||
| O 4HOH/D | 2.44 | 1.0 | 19.5 | |||||
| 1Ca/F1 | Ca2+ | 0.2 | 19.1 | |||||
| 1Ca/G1 | Ca2+ | 0.6 | 13.1 | 2.0 | ||||
| OP1 7G/A | 2.35 | 1.0 | 15.4 | 18.7 | ||||
| OP1 7G/A d | 2.35 | 1.0 | 15.4 | 18.7 | ||||
| OP1 7G/A d | 2.35 | 1.0 | 15.4 | 18.7 | ||||
| O 41HOH/D | 2.39 | 1.0 | 19.1 | |||||
| 41HOH/D d | 2.39 | 1.0 | 19.1 | |||||
| 41HOH/D d | 2.39 | 1.0 | 19.1 | |||||
| 1Cu/C4 | Cu2+ | 0.2 | 25.1 | - | ||||
| O 49HOH/D | 2.19 | 1.0 | 30.6 | N7 8A/B | 1.4 | |||
| O 47HOH/D | 2.22 | 1.0 | 34.2 | N6 8A/B | 1.2 | |||
| O 85HOH/D | 2.16 | 0.5 | 33.1 | |||||
| O 86HOH/D | 2.15 | 0.4 | 32.1 | |||||
| O 1HOH/E | 2.21 | 1.0 | 36.1 | |||||
| O 70HOH/D d | 2.11 | 1.00 | 36.6 | |||||
| 1Co/E1 | Co2+ | 0.5 | 8.5 | 2.1 | ||||
| O4 4U/A | 2.42 | 1.0 | 14.4 | 3.9 | ||||
| O 1HOH/C | 2.13 | 1.0 | 12.8 | O6 3G/B | 11.1 | |||
| O 13HOH/C | 2.11 | 1.0 | 15.9 | O6 3G/A | 11.1 | |||
| O 4HOH/C | 2.10 | 1.0 | 15.5 | |||||
| O 6HOH/C | 2.11 | 1.0 | 20.6 | N7 3G/A | 10.2 | |||
| O 5HOH/C | 2.13 | 1.0 | 17.5 | |||||
| O 3HOH/C | 2.10 | 1.0 | 20.4 | |||||
| 1Co/D2 | Co2+ | 0.6 | 12.0 | 1.8 | ||||
| O 1HOH/C | 2.11 | 1.0 | 18.9 | |||||
| O 1HOH/C d | 2.11 | 1.0 | 18.9 | |||||
| O 1HOH/C d | 2.11 | 1.0 | 18.9 | |||||
| OP1 7G/A | 2.15 | 1.0 | 15.3 | 2.6 | ||||
| OP1 7G/A d | 2.15 | 1.0 | 15.3 | 2.6 | ||||
| OP1 7G/A d | 2.15 | 1.0 | 15.3 | 2.6 | ||||
| 1Co/E2 | Co2+ | 0.3 | 12.5 | |||||
| O 81HOH/C | 2.10 | 1.0 | 23.7 | N7 7G/A | 10.2 | |||
| O 37HOH/C | 2.07 | 1.0 | 25.3 | |||||
| O 17HOH/C | 2.12 | 1.0 | 20.3 | OP2 6C/A | 5.1 | |||
| O 61HOH/C | 2.11 | 1.0 | 26.0 | |||||
| O 63HOH/C | 2.10 | 1.0 | 27.6 | |||||
| O 62HOH/C | 2.09 | 1.0 | 32.4 | |||||
| 1Mn/C1 | Mn2+ | 0.7 | 14.7 | 2.1 | ||||
| O 1HOH/F | 2.14 | 1.0 | 20.4 | |||||
| O 2HOH/F | 2.18 | 1.0 | 19.5 | O4 4U/A | 3.3 | |||
| O 3HOH/F | 2.08 | 1.0 | 16.5 | O6 3G/B | 9.3 | |||
| O 15HOH/F | 2.19 | 1.0 | 21.4 | N7 3G/B | 8.0 | |||
| O 16HOH/F | 2.31 | 1.0 | 18.4 | |||||
| O 17HOH/F | 2.21 | 1.0 | 22.9 | |||||
| 1Mn/D1 | Mn2+ | 0.6 | 14.0 | 2.5 | ||||
| O 7HOH/F | 2.17 | 1.0 | 18.9 | O6 7G/A | 9.3 | |||
| O 8HOH/F | 2.21 | 1.0 | 23.2 | |||||
| O 9HOH/F | 2.00 | 1.0 | 22.2 | |||||
| O 10HOH/F | 2.08 | 1.0 | 25.5 | |||||
| O 11HOH/F | 2.05 | 1.0 | 23.4 | |||||
| O 12HOH/F | 2.18 | 1.0 | 18.9 | N7 7G/A | 8.0 | |||
| 1Mn/E1 | Mn2+ | 0.4 | 15.3 | |||||
| O 13HOH/F | 2.10 | 1.0 | 25.9 | |||||
| O 14HOH/F | 2.07 | 1.0 | 23.1 | O6 7G/B | 9.3 | |||
| O 44HOH/F | 2.13 | 1.0 | 26.2 | N7 7G/B | 8.0 | |||
| O 45HOH/F | 2.06 | 1.0 | 30.6 | |||||
| O 37HOH/F d | 2.32 | 1.0 | 26.5 | |||||
| O 43HOH/F d | 2.23 | 1.0 | 26.2 | |||||
| 1Mn/G1 | Mn2+ | 0.7 | 22.2 | |||||
| OP1 7G/A | 2.59 | 1.0 | 28.4 | 4.6 | ||||
| OP1 7G/A d | 2.99 | 1.0 | 28.4 | 4.6 | ||||
| OP1 7G/A d | 2.27 | 1.0 | 28.4 | 4.6 | ||||
| 1Sr/C2 | Sr2+ | 0.6 | 28.5 | 1.4 | ||||
| O 1HOH/N | 2.52 | 1.0 | 33.6 | O4 4U/A | 5.4 | |||
| O 2HOH/Q | 2.61 | 1.0 | 42.5 | |||||
| O 3HOH/D | 2.49 | 1.0 | 36.3 | O6 3G/B | 5.2 | |||
| O 4HOH/D | 2.53 | 1.0 | 30.3 | O6 3G/A | 5.2 | |||
| O 6HOH/D | 2.61 | 1.0 | 35.92 | |||||
| O 7HOH/D | 2.62 | 1.0 | 38.4 | |||||
| 1Sr/F1 | Sr2+ | 0.5 | 35.3 | |||||
| OP1 7G/A | 3.20 | 1.0 | 33.9 | |||||
| OP1 7G/A d | 3.20 | 1.0 | 33.9 | |||||
| OP1 7G/A d | 3.20 | 1.0 | 33.9 | |||||
| 1Tb/1 | Tb3+ | 0.4 | 39.4 | |||||
| OP2 2C/A | 2.53 | 1.0 | 40.9 | - | ||||
| OP2 2C/B | 2.50 | 1.0 | 41.6 | - | ||||
| O 1HOH/ | 2.34 | 1.0 | 45.0 | |||||
| O 14HOH/D | 2.30 | 1.0 | 45.1 | |||||
| O 19HOH/D | 2.55 | 1.0 | 46.6 | O5′/1 U/A | 7.1 | |||
| 1Tb/E2 | Tb3+ | 0.2 | 44.4 | |||||
| OP1 7G/A | 1.0 | 41.8 | - | |||||
| OP1 7G/A d | 1.0 | 41.8 | ||||||
| OP1 7G/A d | 1.0 | 41.8 |
| Ca2+ | Mn2+ | Co2+ | Cu2+ | Sr2+ | Tb3+ | |
|---|---|---|---|---|---|---|
| PDB Code | 4U3L | 4U3O | 4U3R | 4U78 | 4U3P | 4U47 |
| Data collection | ||||||
| λ (Å) | 1.60810 | 1.60000 | 1.60000 | 1.37478 | 1.60000 | 1.60000 |
| Exposure period (s) | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Oscillation range (°) | 0.25 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| Space group | H3 | H3 | H3 | H3 | H3 | H3 |
| Unit cell parameters | ||||||
| a (Å) | 46.88 | 46.33 | 46.97 | 46.81 | 45.48 | 46.51 |
| b (Å) | 46.88 | 46.33 | 46.97 | 46.81 | 45.48 | 46.51 |
| c (Å) | 53.15 | 58.08 | 53.39 | 53.38 | 57.98 | 56.63 |
| Resolution range (Å) | 32.00–1.68 | 23.16–1.8 | 32.34–1.72 | 32.28–1.50 | 32.58–1.87 | 32.82–1.95 |
| Number of reflections | ||||||
| Total | 69,346 | 78,548 | 35,809 | 112,323 | 35,169 | 31,751 |
| Unique | 4726 | 4140 | 4265 | 6864 | 3728 | 3225 |
| Completeness (%) a | 95.2 (77.60) | 95.74 (79.91) | 96.75 (84.68) | 98.39 (90.32) | 99.79 (97.96) | 96.82 (89.57) |
| (I)/(σ(I)) a | 58.00 (16.2) | 43.26 (12.06) | 14.60 (12.22) | 48.32 (4.56) | 34.84 (4.06) | 33.19 (5.35) |
| Average multiplicity | 14.7(4.9) | 19.0(11.4) | 8.4 (2.0) | 16.4 (5.0) | 9.4 (7.0) | 9.8 (9.0) |
| Rmeas a | 0.036 (0.076) | 0.059 (0.23) | 0.042 (0.044) | 0.041 (0.042) | 0.037 (0.48) | 0.052 (0.44) |
| CC1/2 | 100 (97.7) | 100 (99.2) | 99.8 (82.4) | 99.9 (98.2) | 99.9 (98.2) | 99.8 (98.6) |
| Refinement | ||||||
| Rwork | 0.187 | 0.165 | 0.165 | 0.177 | 0.180 | 0.172 |
| Rfree | 0.212 | 0.175 | 0.198 | 0.206 | 0.229 | 0.207 |
| Root mean square deviations (r.m.s.d.) from target values | ||||||
| Bond lengths (Å) | 0.005 | 0.005 | 0.002 | 0.002 | 0.004 | 0.004 |
| Bond angle (Å) | 0.890 | 0.910 | 0.400 | 0.400 | 0.740 | 0.780 |
| Average B-factors (Å2) | ||||||
| Ligands | 24.5 | 16.6 | 10.9 | 20.3 | 32.4 | 41.8 |
| RNA | 22.9 | 21.4 | 15.1 | 18.1 | 28.7 | 39.0 |
| Solvent | 27.1 | 26.1 | 25.9 | 28.5 | 34.2 | 43.9 |
| Number of RNA atoms b | 334 | 334 | 334 | 334 | 334 | 334 |
| Number of solvent molecules | 29 | 65 | 87 | 78 | 64 | 37 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schaffer, M.F.; Peng, G.; Spingler, B.; Schnabl, J.; Wang, M.; Olieric, V.; Sigel, R.K.O. The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations. Int. J. Mol. Sci. 2016, 17, 988. https://doi.org/10.3390/ijms17070988
Schaffer MF, Peng G, Spingler B, Schnabl J, Wang M, Olieric V, Sigel RKO. The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations. International Journal of Molecular Sciences. 2016; 17(7):988. https://doi.org/10.3390/ijms17070988
Chicago/Turabian StyleSchaffer, Michelle F., Guanya Peng, Bernhard Spingler, Joachim Schnabl, Meitian Wang, Vincent Olieric, and Roland K. O. Sigel. 2016. "The X-ray Structures of Six Octameric RNA Duplexes in the Presence of Different Di- and Trivalent Cations" International Journal of Molecular Sciences 17, no. 7: 988. https://doi.org/10.3390/ijms17070988