High Expression of XRCC6 Promotes Human Osteosarcoma Cell Proliferation through the β-Catenin/Wnt Signaling Pathway and Is Associated with Poor Prognosis
Abstract
:1. Introduction
2. Results
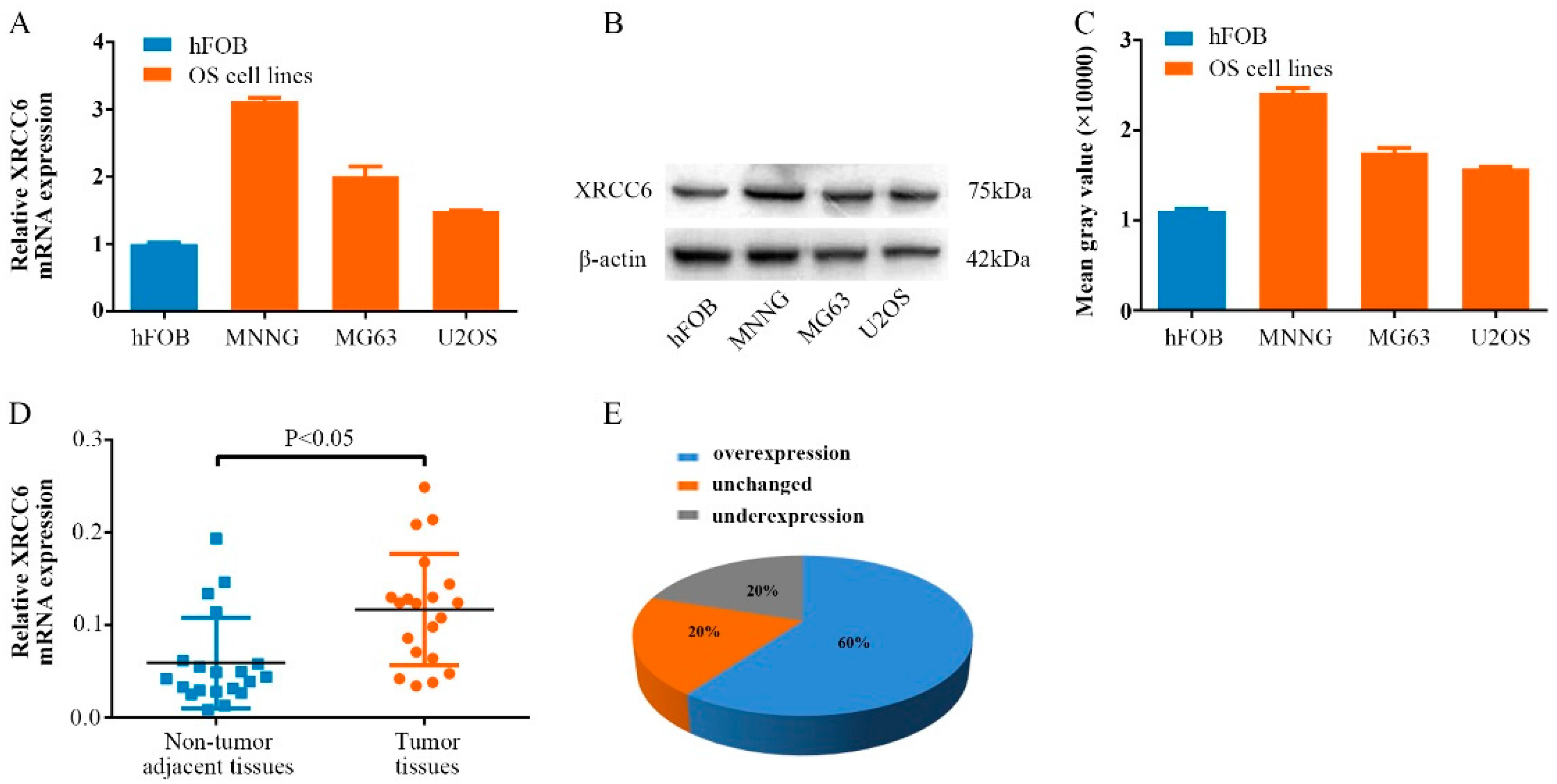
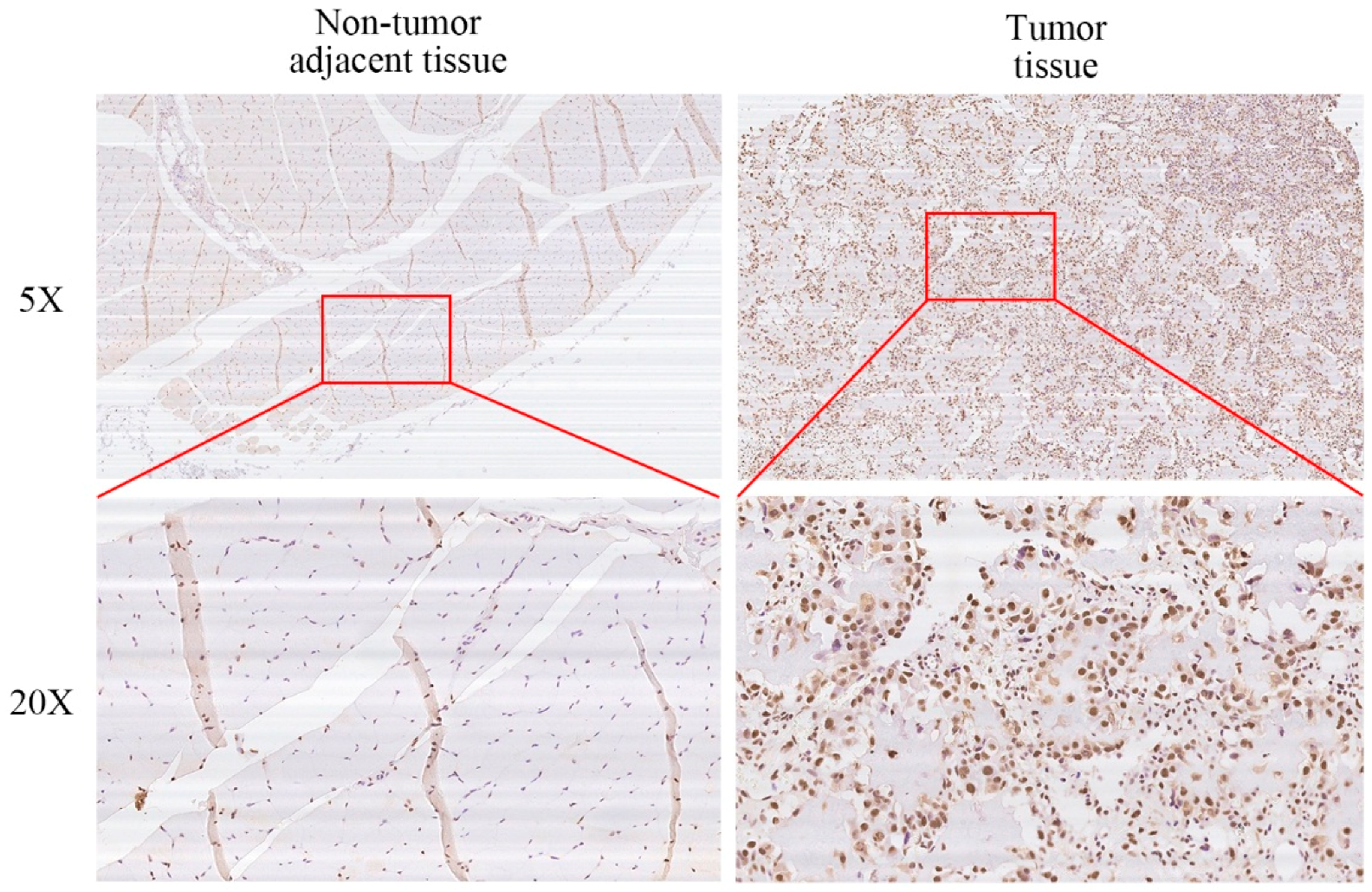
2.1. XRCC6 Is Overexpressed in Human Osteosarcoma (OS) Samples and Cell Lines
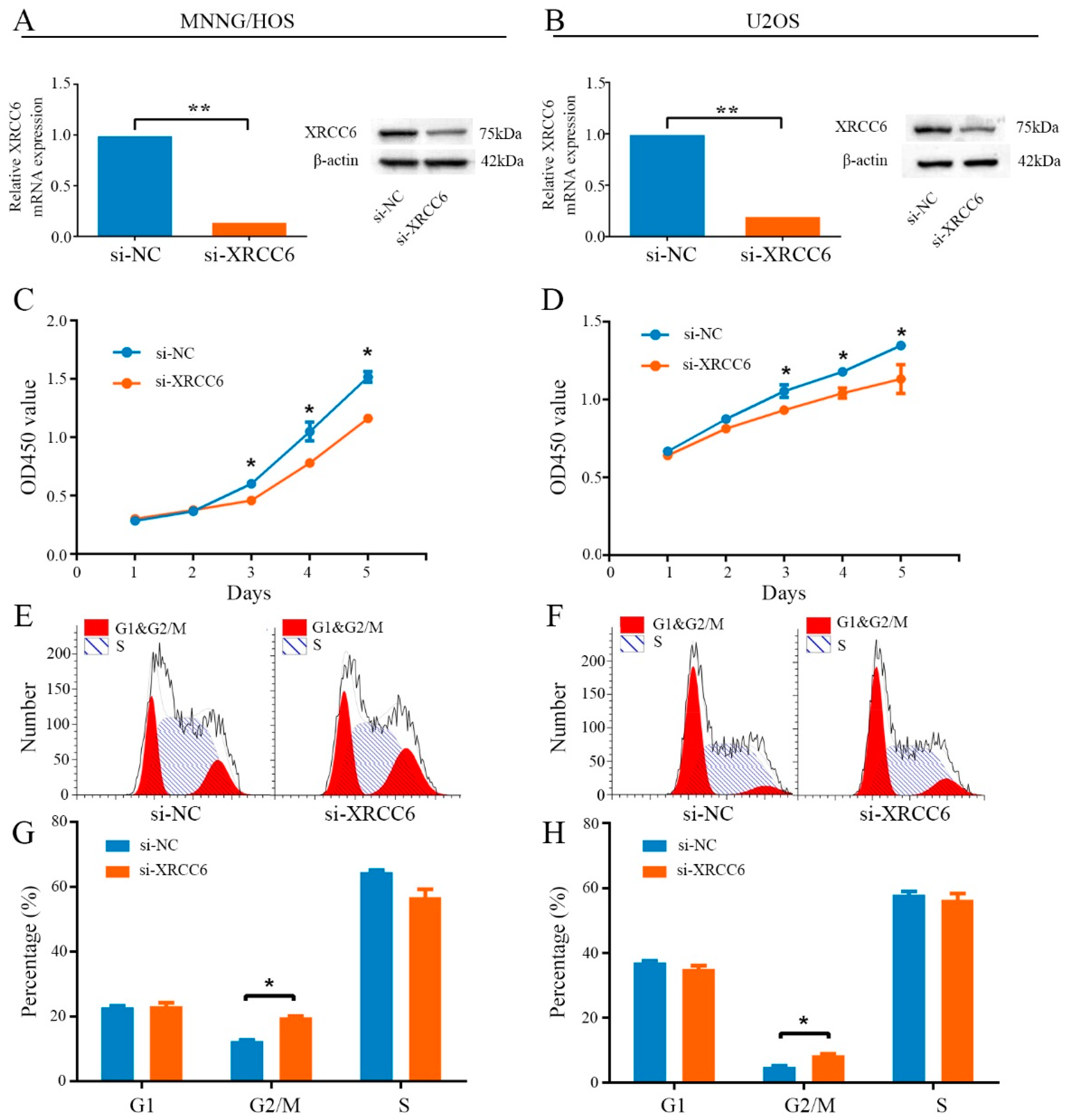
2.2. Knockdown of XRCC6 Expression Inhibited OS Cell Proliferation through G2/M Phase Arrest in Vitro
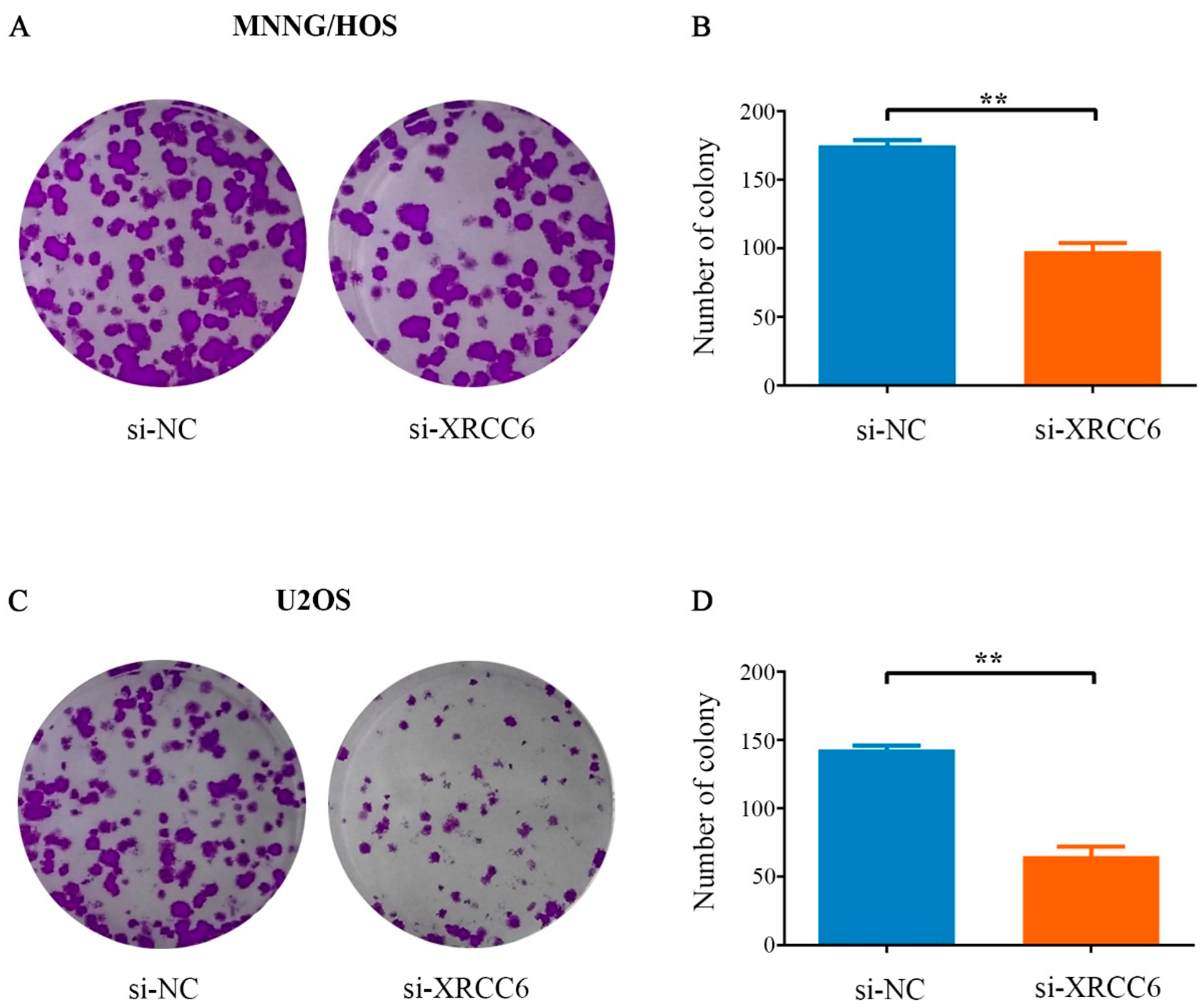
2.3. Decreased XRCC6 Expression Impaired Colony-Forming Capacity of OS Cells
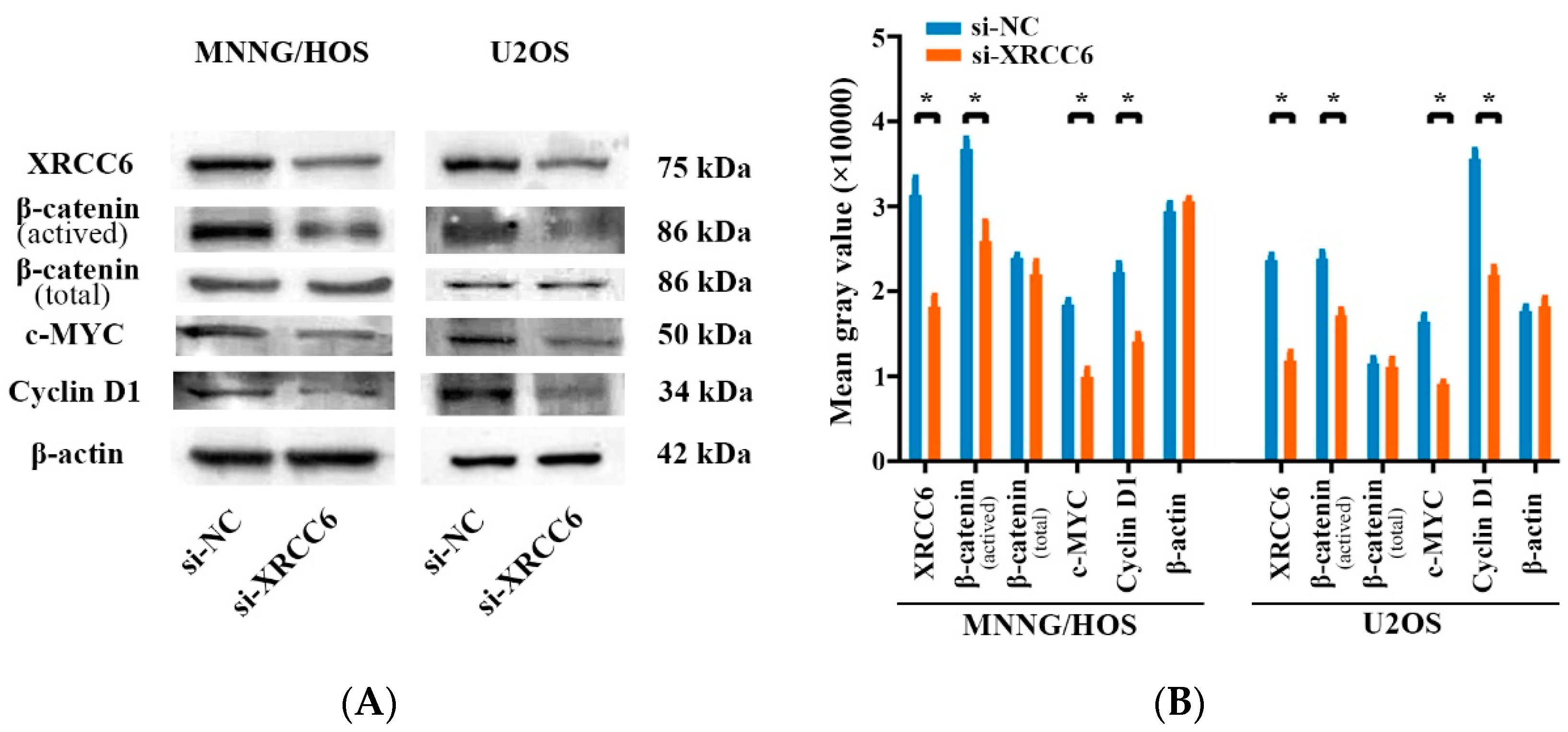
2.4. The β-Catenin/Wnt Signaling Pathway Was Dysregulated by XRCC6 in OS Cells
2.5. XRCC6 Expression Correlates to OS Clinical Stage and Tumor Size
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Cell Culture
4.2. Human Osteosarcoma Samples
4.3. RNA Isolation and qRT-PCR Assays
4.4. Cell Transfection
4.5. Cell Proliferation and Cell Cycle Assay
4.6. Migration and Invasion Assays
4.7. Colony Formation Assay
4.8. Western Blot Analysis
4.9. Immunohistochemistry (IHC)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Moore, D.D.; Luu, H.H. Osteosarcoma. Cancer Treat. Res. 2014, 162, 65–92. [Google Scholar] [PubMed]
- Saha, D.; Saha, K.; Banerjee, A.; Jash, D. Osteosarcoma relapse as pleural metastasis. South Asian J. Cancer 2013, 2, 56. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, A.B.; Li, F.; Li, M.; He, B.; He, X.; Chen, G.; Guo, B.; Li, D.; Jiang, F.; Dang, L. Present Advances and Future Perspectives of Molecular Targeted Therapy for Osteosarcoma. Int. J. Mol. Sci. 2016, 17, 506. [Google Scholar] [CrossRef] [PubMed]
- Coventry, M.B.; Dahlin, D.C. Osteogenic sarcoma; a critical analysis of 430 cases. J. Bone Jt. Surg. Am. Vol. 1957, 39, 741–757. [Google Scholar]
- Allison, D.C.; Carney, S.C.; Ahlmann, E.R.; Hendifar, A.; Chawla, S.; Fedenko, A.; Angeles, C.; Menendez, L.R. A meta-analysis of osteosarcoma outcomes in the modern medical era. Sarcoma 2012, 2012, 704872. [Google Scholar] [CrossRef] [PubMed]
- Bielack, S.S.; Kempf-Bielack, B.; Delling, G.; Exner, G.U.; Flege, S.; Helmke, K.; Kotz, R.; Salzer-Kuntschik, M.; Werner, M.; Winkelmann, W. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: An analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. 2002, 20, 776–790. [Google Scholar] [CrossRef]
- Yang, J.; Cheng, D.; Zhu, B.; Zhou, S.; Ying, T.; Yang, Q. Chromobox Homolog 4 is Positively Correlated to Tumor Growth, Survival and Activation of HIF-1alpha Signaling in Human Osteosarcoma under Normoxic Condition. J. Cancer 2016, 7, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Yin, X.; Yang, H.; Yang, J.; Zhao, J.; Xu, C.; Xu, H. Association of the genetic polymorphisms in XRCC6 and XRCC5 with the risk of ESCC in a high-incidence region of North China. Tumori 2015, 101, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Rathaus, M.; Lerrer, B.; Cohen, H.Y. DeubiKuitylation: A novel DUB enzymatic activity for the DNA repair protein, Ku70. Cell Cycle 2009, 8, 1843–1852. [Google Scholar] [CrossRef] [PubMed]
- Kragelund, B.B.; Weterings, E.; Hartmann-Petersen, R.; Keijzers, G. The Ku70/80 ring in Non-Homologous End-Joining: Easy to slip on, hard to remove. Front. Biosci. 2016, 21, 514–527. [Google Scholar]
- Hsia, T.C.; Liu, C.J.; Chu, C.C.; Hang, L.W.; Chang, W.S.; Tsai, C.W.; Wu, C.I.; Lien, C.S.; Liao, W.L.; Ho, C.Y. Association of DNA double-strand break gene XRCC6 genotypes and lung cancer in Taiwan. Anticancer Res. 2012, 32, 1015–1020. [Google Scholar] [PubMed]
- Zhao, P.; Zou, P.; Zhao, L.; Yan, W.; Kang, C.; Jiang, T.; You, Y. Genetic polymorphisms of DNA double-strand break repair pathway genes and glioma susceptibility. BioMed Cent. Cancer 2013, 13, 234. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Tang, H.; Yu, J.; Zhuang, D.; Zhang, H. Blood-based DNA methylation of DNA repair genes in the non-homologous end-joining (NEHJ) pathway in patient with glioma. Int. J. Clin. Exp. Pathol. 2015, 8, 9463–9467. [Google Scholar] [PubMed]
- Hsu, C.M.; Yang, M.D.; Chang, W.S.; Jeng, L.B.; Lee, M.H.; Lu, M.C.; Chang, S.C.; Tsai, C.W.; Tsai, Y.; Tsai, F.J. The contribution of XRCC6/Ku70 to hepatocellular carcinoma in Taiwan. Anticancer Res. 2013, 33, 529–535. [Google Scholar] [PubMed]
- Wang, Z.; Lin, H.; Hua, F.; Hu, Z.W. Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy 2013, 9, 925–927. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhu, B.; Liu, X.; Yu, H.; Yong, L.; Liu, X.; Shao, J.; Liu, Z. Inhibition of oleandrin on the proliferation show and invasion of osteosarcoma cells in vitro by suppressing Wnt/β-catenin signaling pathway. J. Exp. Clin. Cancer Res. CR 2015, 34, 115. [Google Scholar] [CrossRef] [PubMed]
- Kansara, M.; Tsang, M.; Kodjabachian, L.; Sims, N.A.; Trivett, M.K.; Ehrich, M.; Dobrovic, A.; Slavin, J.; Choong, P.F.; Simmons, P.J. Wnt inhibitory factor 1 is epigenetically silenced in human osteosarcoma, and targeted disruption accelerates osteosarcomagenesis in mice. J. Clin. Investig. 2009, 119, 837–851. [Google Scholar] [CrossRef] [PubMed]
- McQueen, P.; Ghaffar, S.; Guo, Y.; Rubin, E.M.; Zi, X.; Hoang, B.H. The Wnt signaling pathway: Implications for therapy in osteosarcoma. Expert Rev. Anticancer Ther. 2011, 11, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ren, Y.; Han, E.Q.; Li, H.; Chen, D.; Jacobs, J.J.; Gitelis, S.; O’Keefe, R.J.; Konttinen, Y.T.; Yin, G. Inhibition of the Wnt/β-catenin and Notch signaling pathways sensitizes osteosarcoma cells to chemotherapy. Biochem. Biophys. Res. Commun. 2013, 431, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Tavana, O.; Puebla-Osorio, N.; Kim, J.; Sang, M.; Jang, S.; Zhu, C. Ku70 functions in addition to nonhomologous end joining in pancreatic β-cells: A connection to β-catenin regulation. Diabetes 2013, 62, 2429–2438. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.W.; Nam, H.Y.; Kim, H.J.; Moon, S.Y.; Kim, M.R.; Lee, M.; Kim, G.C.; Kim, S.W.; Kim, S.Y. Effect of β-catenin silencing in overcoming radioresistance of head and neck cancer cells by antagonizing the effects of AMPK on Ku70/Ku80. Head Neck 2016, 38, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Guo, Q.; Wen, J.; Li, C.; Lin, J.; Cui, X.; Sang, N.; Pan, J. Survival analyses correlate stanniocalcin 2 overexpression to poor prognosis of nasopharyngeal carcinomas. J. Exp. Clin. Cancer Res.: CR 2014, 33, 26. [Google Scholar] [CrossRef] [PubMed]
- Raymond, A.K.; Jaffe, N. Osteosarcoma multidisciplinary approach to the management from the pathologist’s perspective. Cancer Treat. Res. 2009, 152, 63–84. [Google Scholar] [PubMed]
- Ando, K.; Heymann, M.F.; Stresing, V.; Mori, K.; Redini, F.; Heymann, D. Current therapeutic strategies and novel approaches in osteosarcoma. Cancers 2013, 5, 591–616. [Google Scholar] [CrossRef] [PubMed]
- Luetke, A.; Meyers, P.A.; Lewis, I.; Juergens, H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat. Rev. 2014, 40, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Marina, N.; Gebhardt, M.; Teot, L.; Gorlick, R. Biology and therapeutic advances for pediatric osteosarcoma. Oncologist 2004, 9, 422–441. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Li, H.; Zou, J.; Mi, J.; Liu, F.; Wang, D.; Yan, D.; Wang, B.; Zhang, S.; Tian, G. The relationship between five non-synonymous polymorphisms within three XRCC genes and gastric cancer risk in a Han Chinese population. Tumour Biol. 2016, 37, 5905–5910. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M. Oxidatively induced DNA damage and its repair in cancer. Mutat. Res. Rev. Mutat. Res. 2015, 763, 212–245. [Google Scholar] [CrossRef] [PubMed]
- Qiao, W.; Wang, T.; Zhang, L.; Tang, Q.; Wang, D.; Sun, H. Association study of single nucleotide polymorphisms in XRCC1 gene with the risk of gastric cancer in Chinese population. Int. J. Biol. Sci. 2013, 9, 753–758. [Google Scholar] [CrossRef] [PubMed]
- Perez, L.O.; Crivaro, A.; Barbisan, G.; Poleri, L.; Golijow, C.D. XRCC2 R188H (rs3218536), XRCC3 T241M (rs861539) and R243H (rs77381814) single nucleotide polymorphisms in cervical cancer risk. Pathol. Oncol. Res. 2013, 19, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Curtin, K.; Lin, W.Y.; George, R.; Katory, M.; Shorto, J.; Cannon-Albright, L.A.; Smith, G.; Bishop, D.T.; Cox, A.; Camp, N.J. Genetic variants in XRCC2: New insights into colorectal cancer tumorigenesis. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2476–2484. [Google Scholar] [CrossRef] [PubMed]
- Moeller, B.J.; Yordy, J.S.; Williams, M.D.; Giri, U.; Raju, U.; Molkentine, D.P.; Byers, L.A.; Heymach, J.V.; Story, M.D.; Lee, J.J. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clin. Cancer Res. 2011, 17, 2035–2043. [Google Scholar] [CrossRef] [PubMed]
- Marimuthu, A.; Chavan, S.; Sathe, G.; Sahasrabuddhe, N.A.; Srikanth, S.M.; Renuse, S.; Ahmad, S.; Radhakrishnan, A.; Barbhuiya, M.A.; Kumar, R.V. Identification of head and neck squamous cell carcinoma biomarker candidates through proteomic analysis of cancer cell secretome. Biochim. Biophys. Acta 2013, 1834, 2308–2316. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Chen, B.S. A network-based biomarker approach for molecular investigation and diagnosis of lung cancer. BioMed Cent. Med. Genom. 2011, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.W.; Kim, H.; Kim, K.H. Expression of Ku70 and Ku80 mediated by NF-κB and cyclooxygenase-2 is related to proliferation of human gastric cancer cells. J. Biol. Chem. 2002, 277, 46093–46100. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, X.; Shi, W.; Xu, J.; Fan, H.; Zhang, S.; Ni, R. The DNA damage repair protein Ku70 regulates tumor cell and hepatic carcinogenesis by interacting with FOXO4. Pathol. Res. Pract. 2016, 212, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Lin, L.; Lama, S.; Qiao, M.; Tuor, U.I. Cerebral expression of DNA repair protein, Ku70, and its association with cell proliferation following cerebral hypoxia-ischemia in neonatal rats. Int. J. Dev. Neurosci. 2009, 27, 129–134. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, B.T.; Tamai, K.; He, X. Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev. Cell 2009, 17, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; He, H.; Lei, G. Wnt/β-catenin pathway in bone cancers. Tumour Biol. 2014, 35, 9439–9445. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Ji, T.; Chen, C.F.; Hoang, B.H. Wnt signaling in osteosarcoma. Adv. Exp. Med. Biol. 2014, 804, 33–45. [Google Scholar] [PubMed]
- Cai, Y.; Cai, T.; Chen, Y. Wnt pathway in osteosarcoma, from oncogenic to therapeutic. J. Cell. Biochem. 2014, 115, 625–631. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.H.; Guo, Y.; Ghaffar, S.; McQueen, P.; Pourmorady, J.; Christ, A.; Rooney, K.; Ji, T.; Eskander, R.; Zi, X. Dkk-3, a secreted wnt antagonist, suppresses tumorigenic potential and pulmonary metastasis in osteosarcoma. Sarcoma 2013, 2013, 147541. [Google Scholar] [CrossRef] [PubMed]
- Puebla-Osorio, N.; Kim, J.; Ojeda, S.; Zhang, H.; Tavana, O.; Li, S.; Wang, Y.; Ma, Q.; Schluns, K.S.; Zhu, C. A novel Ku70 function in colorectal homeostasis separate from nonhomologous end joining. Oncogene 2014, 33, 2748–2757. [Google Scholar] [CrossRef] [PubMed]
- Idogawa, M.; Masutani, M.; Shitashige, M.; Honda, K.; Tokino, T.; Shinomura, Y.; Imai, K.; Hirohashi, S.; Yamada, T. Ku70 and poly(ADP-ribose) polymerase-1 competitively regulate β-catenin and T-cell factor-4-mediated gene transactivation: Possible linkage of DNA damage recognition and Wnt signaling. Cancer Res. 2007, 67, 911–918. [Google Scholar] [CrossRef] [PubMed]
| Clinicopathologic Parameters | Number of Cases | IHC of XRCC6 | |||
|---|---|---|---|---|---|
| Positive | Negative | χ2 | p Value | ||
| Age (years) | |||||
| <20 | 31 | 17 | 14 | 0.023 | 0.879 |
| ≥20 | 19 | 10 | 9 | ||
| Gender | |||||
| Male | 27 | 14 | 13 | 0.109 | 0.741 |
| Female | 23 | 13 | 10 | ||
| Anatomic Location | |||||
| Tibia/Femur | 33 | 17 | 16 | 0.241 | 0.623 |
| Elsewhere | 17 | 10 | 7 | ||
| Ennecking Stage | |||||
| I/II | 35 | 15 | 20 | 4.432 | 0.035 * |
| III | 15 | 12 | 3 | ||
| Tumor Size (cm3) | |||||
| <50 | 21 | 7 | 14 | 6.226 | 0.013 * |
| ≥50 | 29 | 20 | 9 | ||
| Degree of Malignancy | |||||
| Low | 23 | 9 | 14 | 3.791 | 0.052 |
| High | 27 | 18 | 9 | ||
| Tumor Necrosis Rate (%) | |||||
| <90 | 27 | 16 | 11 | 0.654 | 0.419 |
| ≥90 | 23 | 11 | 12 | ||
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, B.; Cheng, D.; Li, S.; Zhou, S.; Yang, Q. High Expression of XRCC6 Promotes Human Osteosarcoma Cell Proliferation through the β-Catenin/Wnt Signaling Pathway and Is Associated with Poor Prognosis. Int. J. Mol. Sci. 2016, 17, 1188. https://doi.org/10.3390/ijms17071188
Zhu B, Cheng D, Li S, Zhou S, Yang Q. High Expression of XRCC6 Promotes Human Osteosarcoma Cell Proliferation through the β-Catenin/Wnt Signaling Pathway and Is Associated with Poor Prognosis. International Journal of Molecular Sciences. 2016; 17(7):1188. https://doi.org/10.3390/ijms17071188
Chicago/Turabian StyleZhu, Bin, Dongdong Cheng, Shijie Li, Shumin Zhou, and Qingcheng Yang. 2016. "High Expression of XRCC6 Promotes Human Osteosarcoma Cell Proliferation through the β-Catenin/Wnt Signaling Pathway and Is Associated with Poor Prognosis" International Journal of Molecular Sciences 17, no. 7: 1188. https://doi.org/10.3390/ijms17071188







