Expression and Characterization of a Novel Glycerophosphodiester Phosphodiesterase from Pyrococcus furiosus DSM 3638 That Possesses Lysophospholipase D Activity
Abstract
:1. Introduction
2. Results
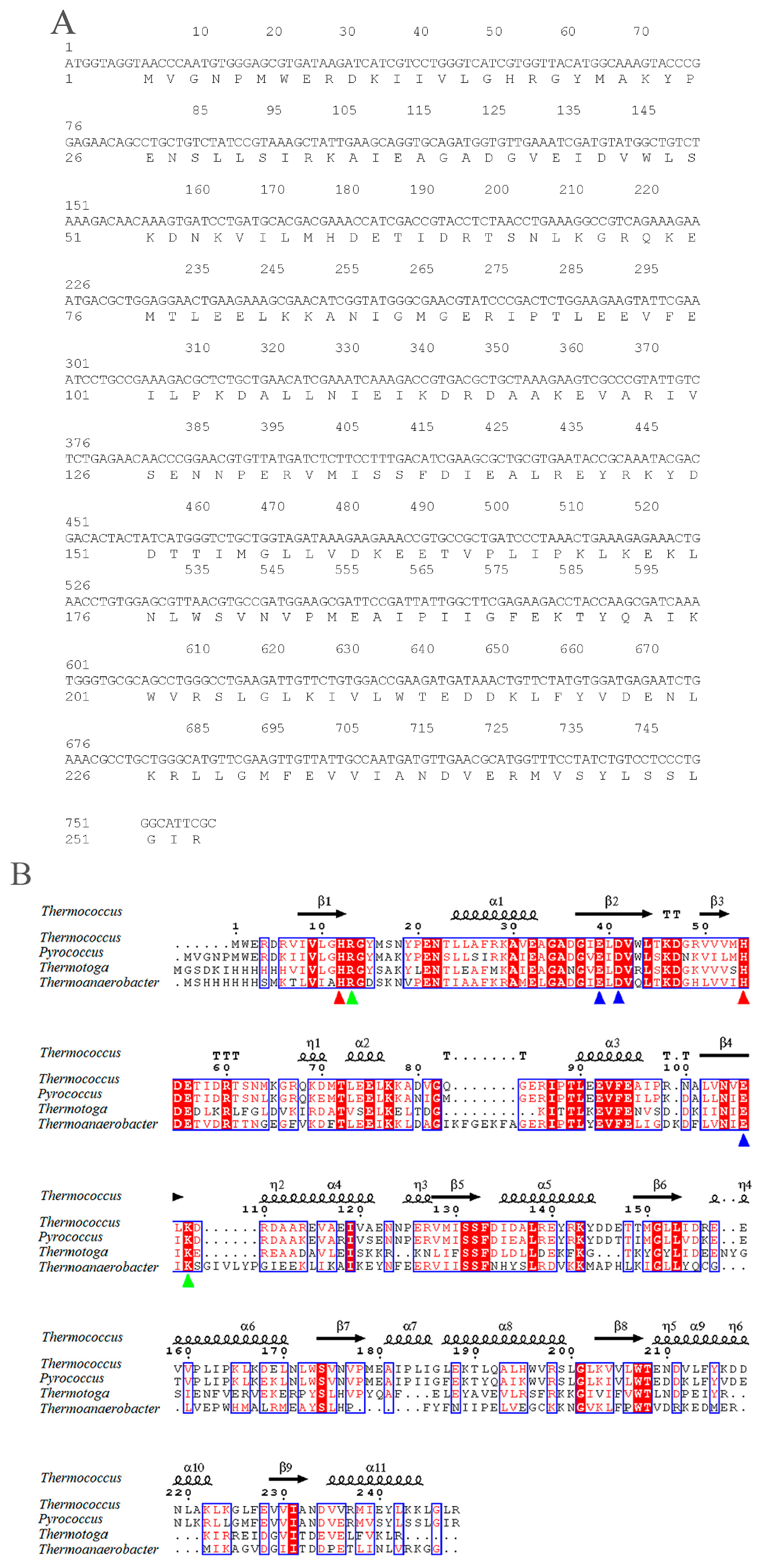
2.1. Sequence Analysis of pfGDPD
2.2. Expression and Purification of the Recombinant pfGDPD
2.3. Biochemical Characterization of Recombinant Enzyme
2.3.1. Effect of Temperature on Enzymatic Activity and Thermostability of Recombinant pfGDPD
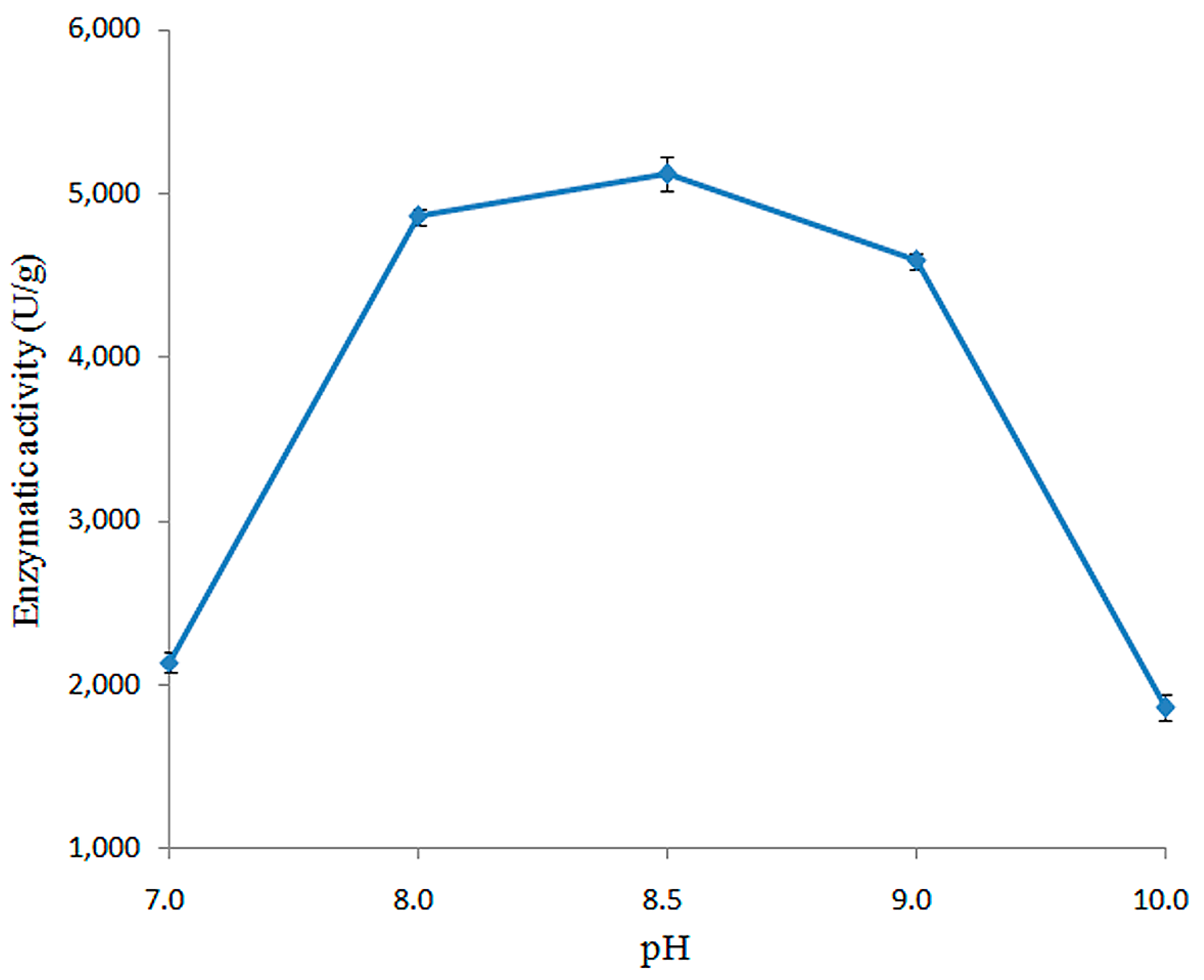
2.3.2. Effect of pH on Enzymatic Activity of Recombined pfGDPD
2.3.3. Effects of Metal Cations on Enzymatic Activity of Recombinant pfGDPD
2.3.4. Effect of Organic Solvents on Recombined pfGDPD Activity
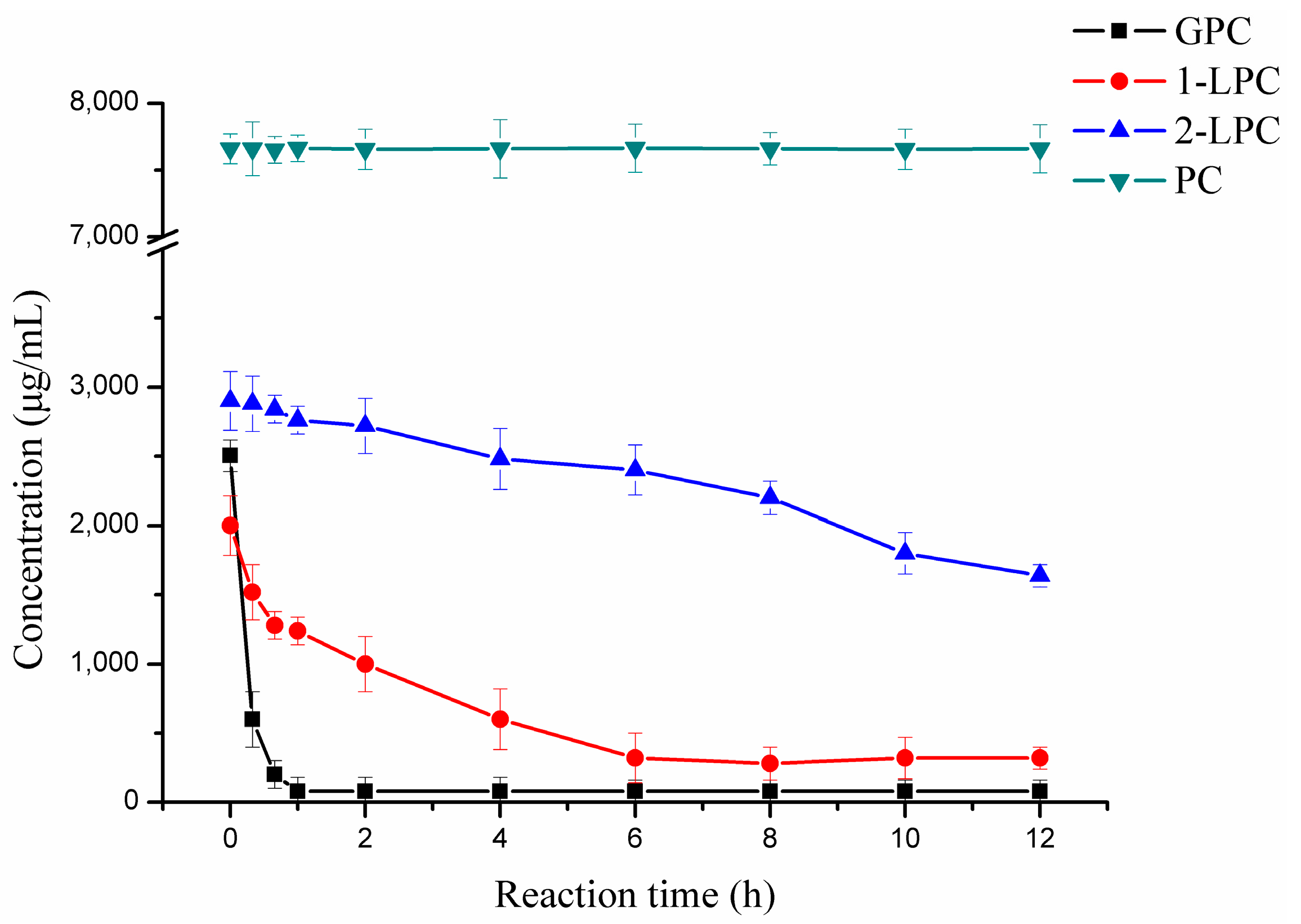
2.4. Hydrolytic Reaction
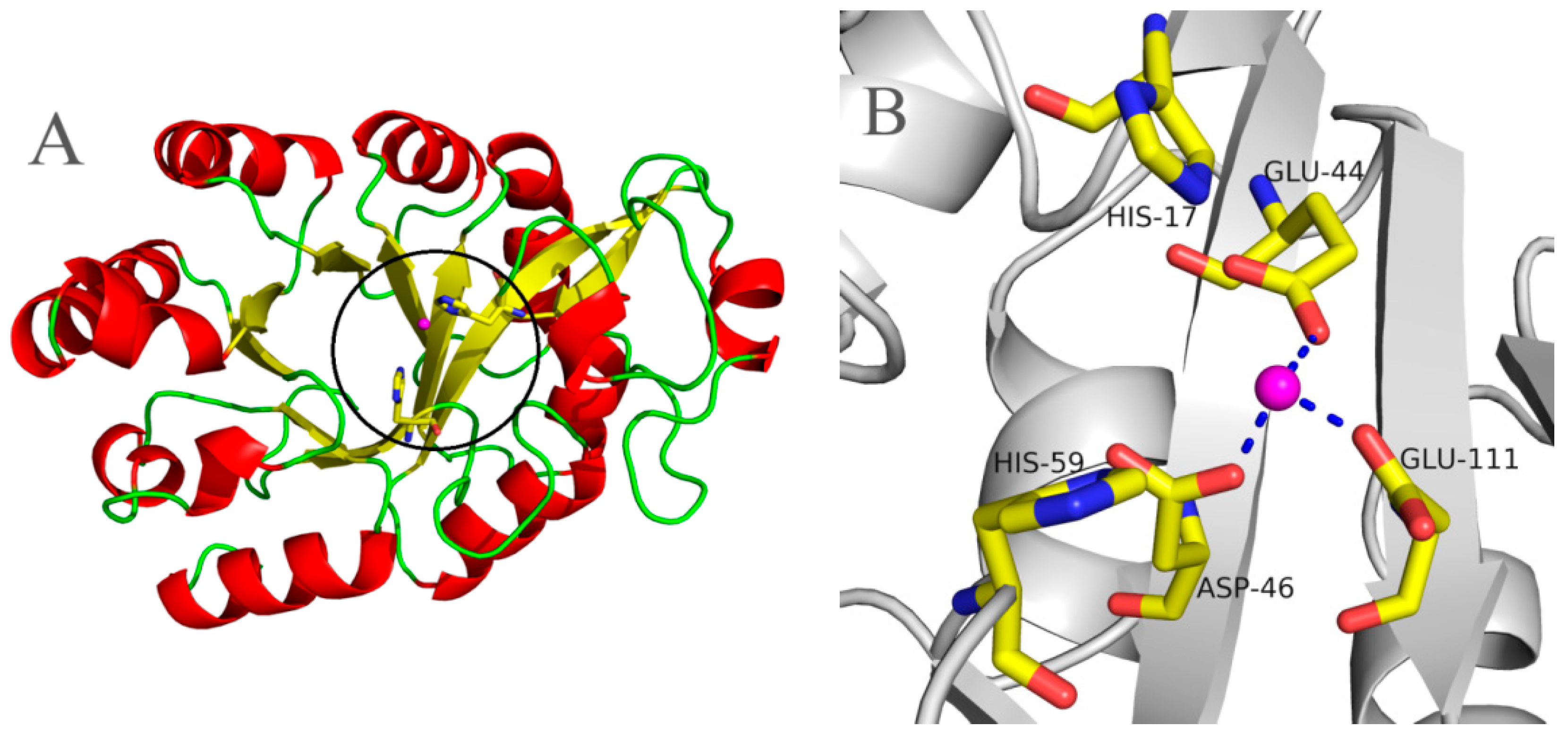
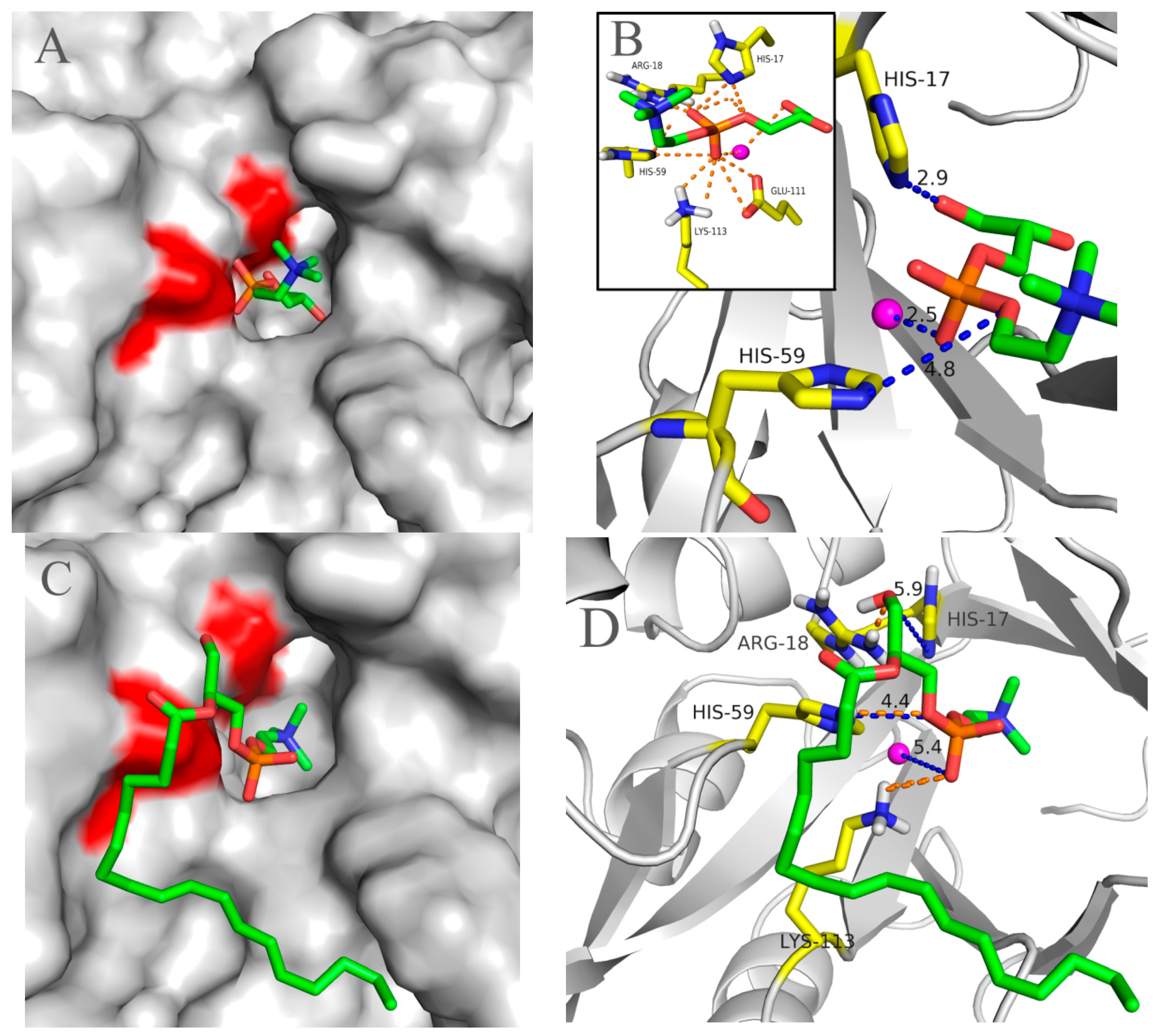
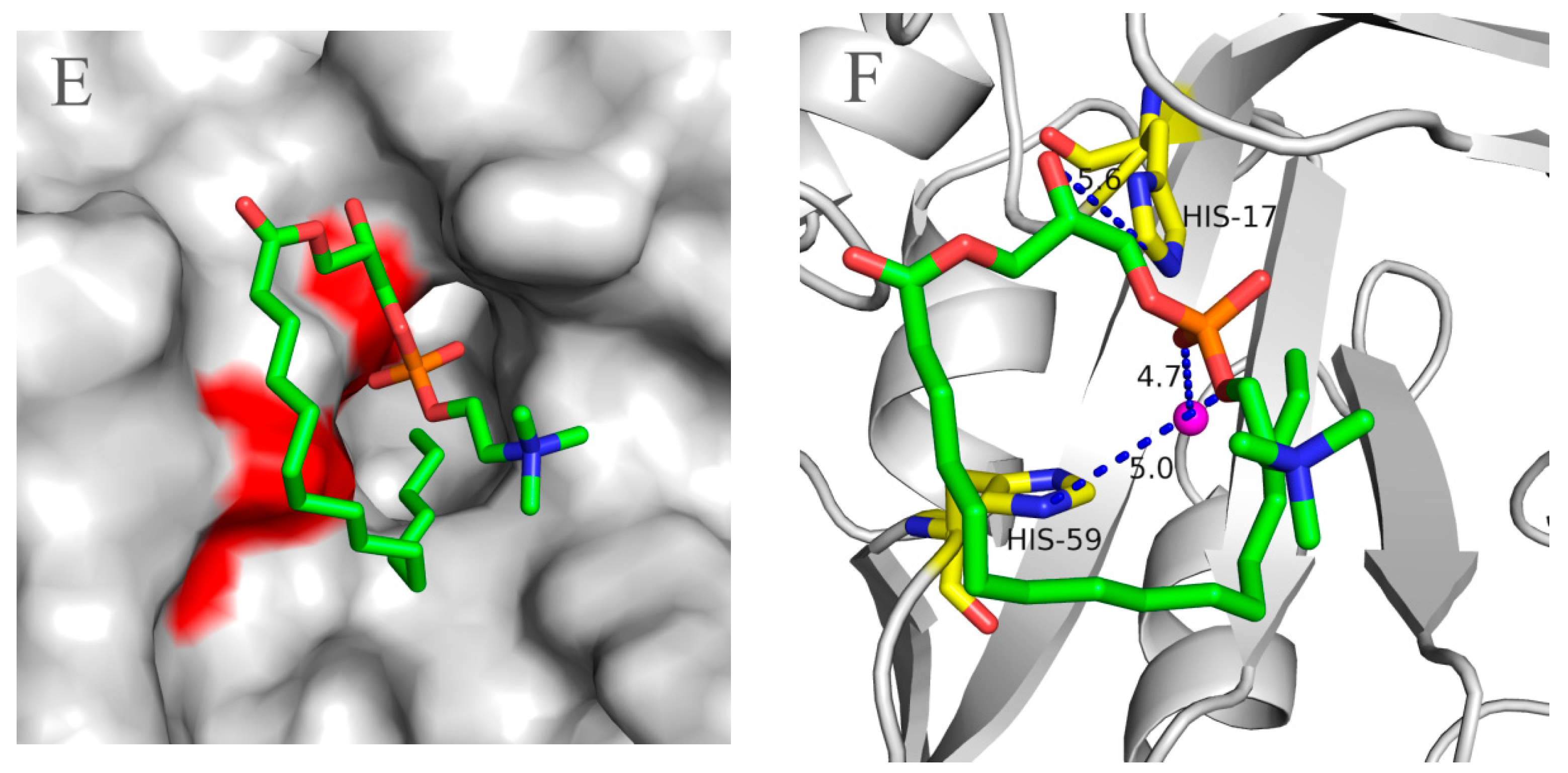
2.5. Molecular 3D-Model of pfGDPD and Docking of GPC, 1-LPC, 2-LPC to pfGDPD
3. Discussion
4. Materials and Methods
4.1. Chemicals, Enzymes and Strains
4.2. Sequence Analysis of pfGDPD
4.3. Expression and Purification of Recombinant pfGDPD
4.4. Biochemical Characterization of Recombinant Enzyme
4.4.1. Enzyme Activity Testing
4.4.2. Determining Temperature Optimum for the Activity and Thermostability of pfGDPD
4.4.3. Determining pH Optimum for the Activity of pfGDPD
4.4.4. Effect of Metal Ions on the Enzymatic Activity of pfGDPD
4.4.5. Effect of Organic Solvents on the Enzymatic Activity of pfGDPD
4.5. Hydrolytic Reaction and Analysis
4.6. Molecular Modeling of pfGDPD and Molecular Docking Simulation
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Raetz, C.R. Molecular genetics of membrane phospholipid synthesis. Annu. Rev. Genet. 1986, 20, 253–295. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Carr, P.D.; Liu, J.W.; Watt, S.J.; Beck, J.L.; Ollis, D.L. The structure and function of a novel glycerophosphodiesterase from Enterobacter aerogenes. J. Mol. Biol. 2007, 367, 1047–1062. [Google Scholar] [CrossRef] [PubMed]
- Larson, T.J.; Ehrmann, M.; Boos, W. Periplasmic glycerophosphodiester phosphodiesterase of Escherichia coli, a new enzyme of the glp regulon. J. Biol. Chem. 1983, 258, 5428–5432. [Google Scholar] [PubMed]
- Brzoska, P.; Boos, W. Characteristics of a ugp-encoded and phoB-dependent glycerophosphoryl diester phosphodiesterase which is physically dependent on the ugp transport system of Escherichia coli. J. Bacteriol. 1988, 170, 4125–4135. [Google Scholar] [PubMed]
- Fan, X.; Goldfine, H.; Lysenko, E.; Weiser, J.N. The transfer of choline from the host to the bacterial cell surface requires GlpQ in Haemophilus influenzae. Mol. Microbiol. 2001, 41, 1029–1036. [Google Scholar] [CrossRef] [PubMed]
- Schmidl, S.R.; Otto, A.; Lluch-Senar, M.; Pinol, J.; Busse, J.; Becher, D.; Stülke, J. A trigger enzyme in Mycoplasma pneumoniae: Impact of the glycerophosphodiesterase GlpQ on virulence and gene expression. PLoS Pathog. 2011, 7, e1002263. [Google Scholar] [CrossRef] [PubMed]
- Yanaka, N. Mammalian glycerophosphodiester phosphodiesterases. Biosci. Biotechnol. Biochem. 2007, 71, 1811–1818. [Google Scholar] [CrossRef] [PubMed]
- Gallazzini, M.; Ferraris, J.D.; Burg, M.B. GDPD5 is a glycerophosphocholine phosphodiesterase that osmotically regulates the osmoprotective organic osmolyte GPC. Proc. Natl. Acad. Sci. USA 2008, 105, 11026–11031. [Google Scholar] [CrossRef] [PubMed]
- Simon, G.M.; Cravatt, B.F. Anandamide biosynthesis catalyzed by the phosphodiesterase GDE1 and detection of glycerophospho-N-acyl ethanolamine precursors in mouse brain. J. Biol. Chem. 2008, 283, 9341–9349. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, Y.; Ohshima, N.; Yoshizawa, I.; Kamei, Y.; Mariggiò, S.; Okamoto, K.; Maeda, M.; Nogusa, Y.; Fujioka, Y.; Izumi, T.; et al. A novel glycerophosphodiester phosphodiesterase, GDE5, controls skeletal muscle development via a non-enzymatic mechanism. J. Biol. Chem. 2010, 285, 27652–27663. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Sockanathan, S. Transmembrane protein GDE2 induces motor neuron differentiation in vivo. Science 2005, 309, 2212–2215. [Google Scholar] [CrossRef] [PubMed]
- Corda, D.; Kudo, T.; Zizza, P.; Iurisci, C.; Kawai, E.; Kato, N.; Yanaka, N.; Mariggiò, S. The developmentally regulated osteoblast phosphodiesterase GDE3 is glycerophosphoinositol-specific and modulates cell growth. J. Biol. Chem. 2009, 284, 24848–24856. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, S.Y.; Jackson, C.; Liu, J.W.; Ollis, D.L. Growth of Escherichia coli coexpressing phosphotriesterase and glycerophosphodiester phosphodiesterase, using paraoxon as the sole phosphorus source. Appl. Environ. Microbiol. 2004, 70, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, E.; Li, Y.; Xu, C.; Raushel, F.M. Characterization of a phosphodiesterase capable of hydrolyzing EA 2192, the most toxic degradation product of the nerve agent VX. Biochemistry 2007, 46, 9032–9040. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.H.; Foo, J.L.; Schenk, G.; Gahan, L.R.; Carr, P.D.; Ollis, D.L. Directed evolution combined with rational design increases activity of GpdQ toward a non-physiological substrate and alters the oligomeric structure of the enzyme. Protein Eng. Des. Sel. 2011, 24, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Daumann, L.J.; Larrabee, J.A.; Ollis, D.; Schenk, G.; Gahan, L.R. Immobilization of the enzyme GpdQ on magnetite nanoparticles for organophosphate pesticide bioremediation. J. Inorg. Biochem. 2014, 131, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitić, N.; Smith, S.J.; Neves, A.; Guddat, L.W.; Gahan, L.R.; Schenk, G. The catalytic mechanisms of binuclear metallohydrolases. Chem. Rev. 2006, 106, 3338–3363. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Liu, J.F.; An, X.M.; Liang, D.C. Crystal structure of glycerophosphodiester phosphodiesterase (GDPD) from Thermoanaerobacter tengcongensis, a metal ion-dependent enzyme: Insight into the catalytic mechanism. Proteins 2008, 72, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Meldrum, E.; Parker, P.J.; Carozzi, A. The PtdIns-PLC superfamily and signal transduction. Biochim. Biophys. Acta 1991, 1092, 49–71. [Google Scholar] [CrossRef]
- Zheng, B.; Berrie, C.P.; Corda, D.; Farquhar, M.G. GDE1/MIR16 is a glycerophosphoinositol phosphodiesterase regulated by stimulation of G protein-coupled receptors. Proc. Natl. Acad. Sci. USA 2003, 100, 1745–1750. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Chen, D.; Farquhar, M.G. MIR16, a putative membrane glycerophosphodiester phosphodiesterase, interacts with RGS16. Proc. Natl. Acad. Sci. USA 2000, 97, 3999–4004. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.J.; Hadler, K.S.; Carr, P.D.; Oakley, A.J.; Yip, S.; Schenk, G.; Ollis, D.L. Malonate-bound structure of the glycerophosphodiesterase from Enterobacter aerogenes (GpdQ) and characterization of the native Fe2+ metal-ion preference. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64 Pt 8, 681–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohshima, N.; Kudo, T.; Yamashita, Y.; Mariggiò, S.; Araki, M.; Honda, A.; Nagano, T.; Isaji, C.; Kato, N.; Corda, D.; et al. New members of the mammalian glycerophosphodiester phosphodiesterase family: GDE4 and GDE7 produce lysophosphatidic acid by lysophospholipase D activity. J. Biol. Chem. 2015, 290, 4260–4271. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, K.; Okamoto, Y.; Rahman, I.A.; Uyama, T.; Inoue, T.; Tokumura, A.; Ueda, N. Glycerophosphodiesterase GDE4 as a novel lysophospholipase D: A possible involvement in bioactive N-acylethanolamine biosynthesis. Biochim. Biophys. Acta 2015, 1851, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Prediction of transmembrane helices in proteins. TMHMM Server v. 2.0. Available online: http://www.cbs.dtu.dk/services/TMHMM-2.0/ (accessed on 15 May 2016).
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, W29–W33. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, T.; Qiao, J.; Liu, X.; Bo, J.; Wang, J.; Lu, F. High-yield phosphatidylserine production via yeast surface display of phospholipase D from Streptomyces chromofuscus on Pichia pastoris. J. Agric. Food Chem. 2014, 62, 5354–5360. [Google Scholar] [CrossRef] [PubMed]
- Lan, D.M.; Wang, Q.; Xu, J.X.; Zhou, P.F.; Yang, B.; Wang, Y.H. Residue Asn277 affects the stability and substrate specificity of the SMG1 lipase from Malassezia globosa. Int. J. Mol. Sci. 2015, 16, 7273–7288. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.P.; Xu, H.; Lan, D.M.; Yang, B.; Wang, Y.H. Hydrolysis of lysophosphatidylcholines by a lipase from Malassezia globosa. Eur. J. Lipid Sci. Technol. 2015, 117, 1655–1658. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Cassarino, T.G.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, W252–W258. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: Palo Alto, CA, USA, 2014. [Google Scholar]




| Ions | Concentration (mM) | Enzymatic Activity (U/g) |
|---|---|---|
| None | — | N.D. |
| EDTA | 10 | N.D. |
| Mn2+ | 1 | 4515 ± 187 |
| 5 | 5842 ± 6 | |
| 10 | 5677 ± 51 | |
| 50 | 2864 ± 129 | |
| 100 | 1967 ± 266 | |
| Co2+ | 1 | 4972 ± 58 |
| 5 | 3935 ± 138 | |
| 10 | 2903 ± 133 | |
| 50 | 1536 ± 229 | |
| 100 | 1128 ± 179 | |
| Ni2+ | 1 | 1041 ± 42 |
| 5 | 1893 ± 54 | |
| 10 | 1723 ± 90 | |
| 50 | 719 ± 93 | |
| 100 | 359 ± 69 | |
| Cu2+ | 10 | 370 ± 202 |
| Mg2+ | 10 | 74 ± 19 |
| Ca2+ | 10 | N.D. |
| Li+ | 10 | N.D. |
| Fe3+ | 10 | N.D. |
| Zn2+ | 10 | N.D. |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Lai, L.; Liu, Y.; Yang, B.; Wang, Y. Expression and Characterization of a Novel Glycerophosphodiester Phosphodiesterase from Pyrococcus furiosus DSM 3638 That Possesses Lysophospholipase D Activity. Int. J. Mol. Sci. 2016, 17, 831. https://doi.org/10.3390/ijms17060831
Wang F, Lai L, Liu Y, Yang B, Wang Y. Expression and Characterization of a Novel Glycerophosphodiester Phosphodiesterase from Pyrococcus furiosus DSM 3638 That Possesses Lysophospholipase D Activity. International Journal of Molecular Sciences. 2016; 17(6):831. https://doi.org/10.3390/ijms17060831
Chicago/Turabian StyleWang, Fanghua, Linhui Lai, Yanhua Liu, Bo Yang, and Yonghua Wang. 2016. "Expression and Characterization of a Novel Glycerophosphodiester Phosphodiesterase from Pyrococcus furiosus DSM 3638 That Possesses Lysophospholipase D Activity" International Journal of Molecular Sciences 17, no. 6: 831. https://doi.org/10.3390/ijms17060831





