A Chloroplast-Localized Rubredoxin Family Protein Gene from Puccinellia tenuiflora (PutRUB) Increases NaCl and NaHCO3 Tolerance by Decreasing H2O2 Accumulation
Abstract
:1. Introduction
2. Results and Discussion
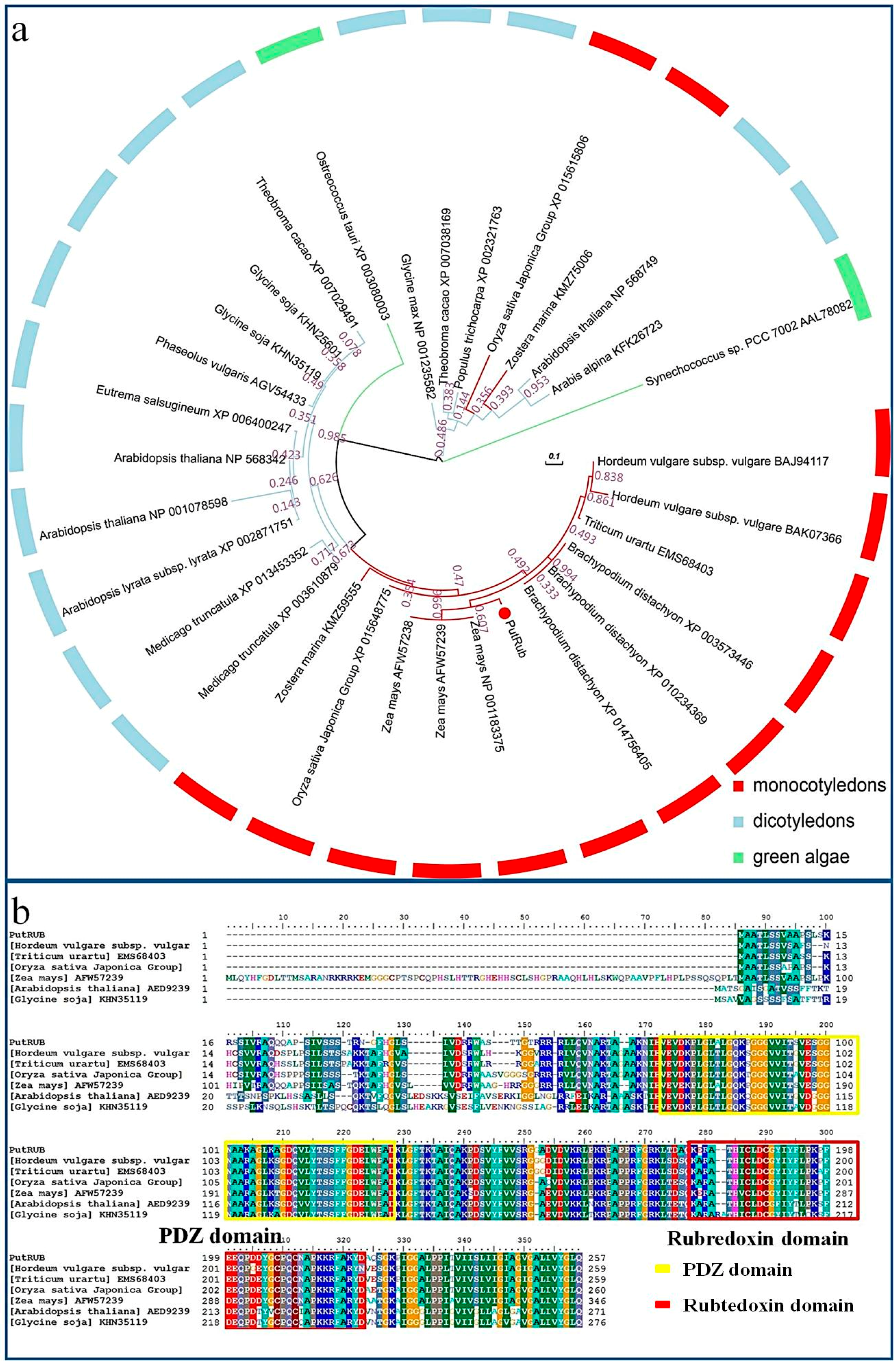
2.1. Characteristics of PutRUB
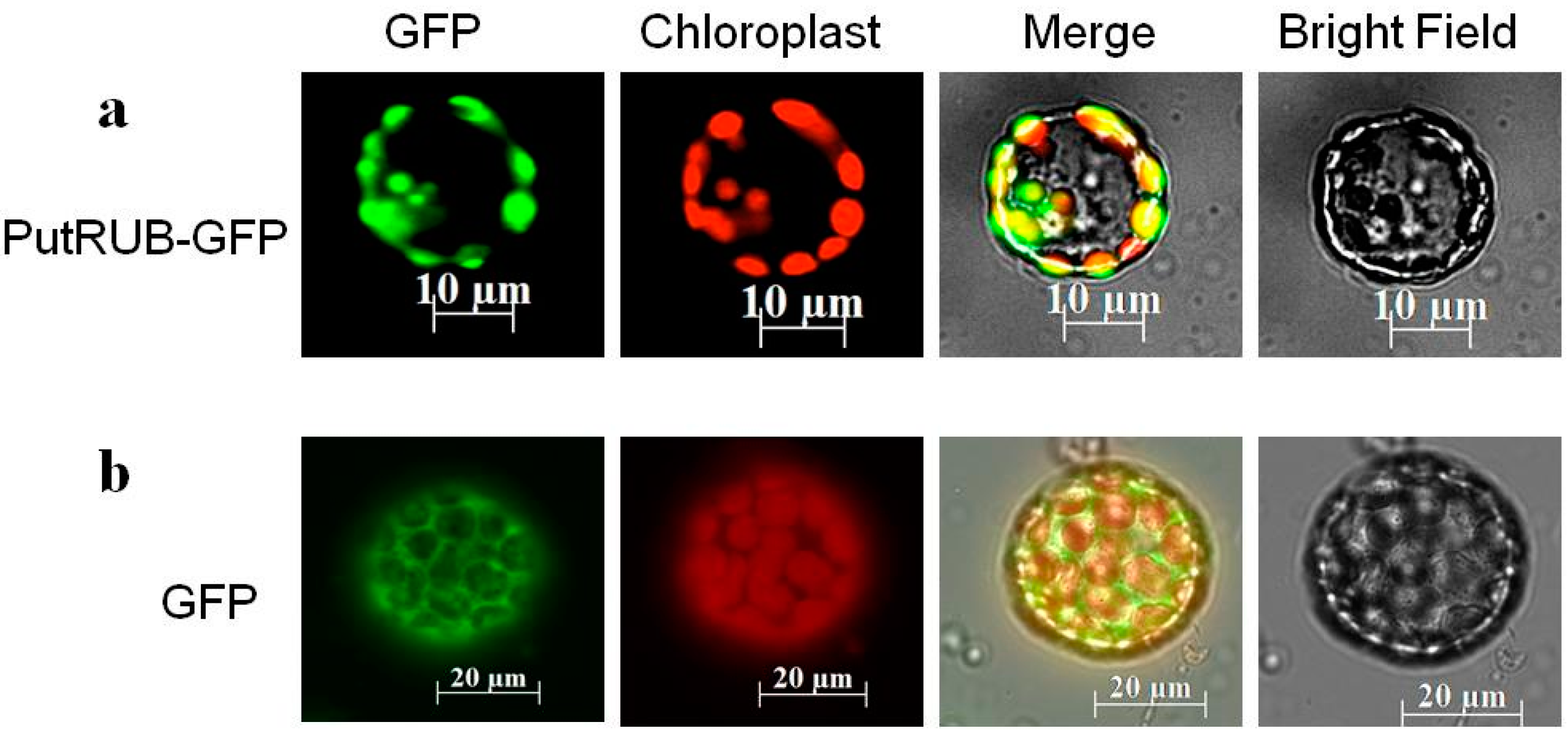
2.2. Subcellular Localization of the PutRUB: GFP Fusion Protein
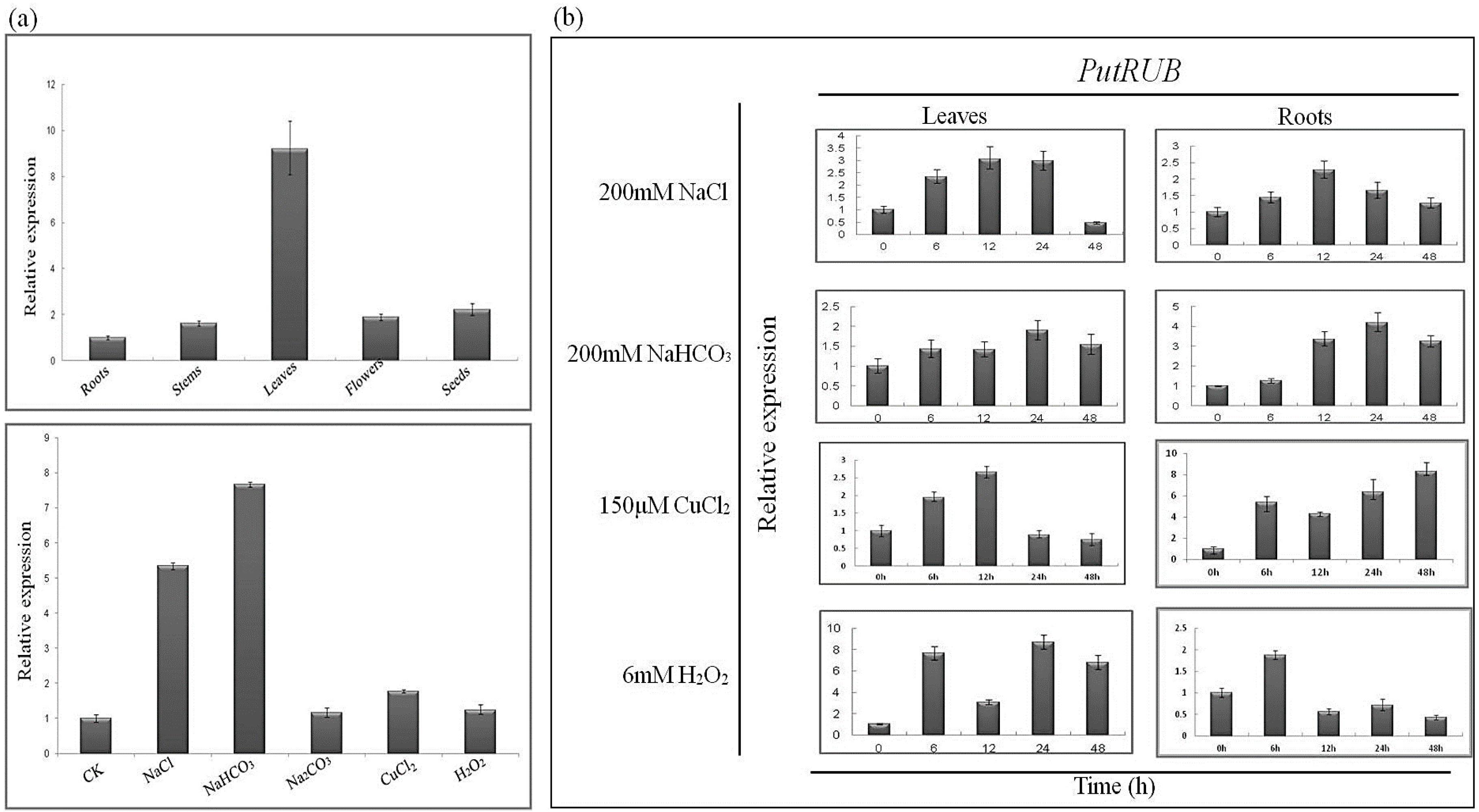
2.3. Expression of the PutRUB Gene Is Induced by Abiotic Stresses
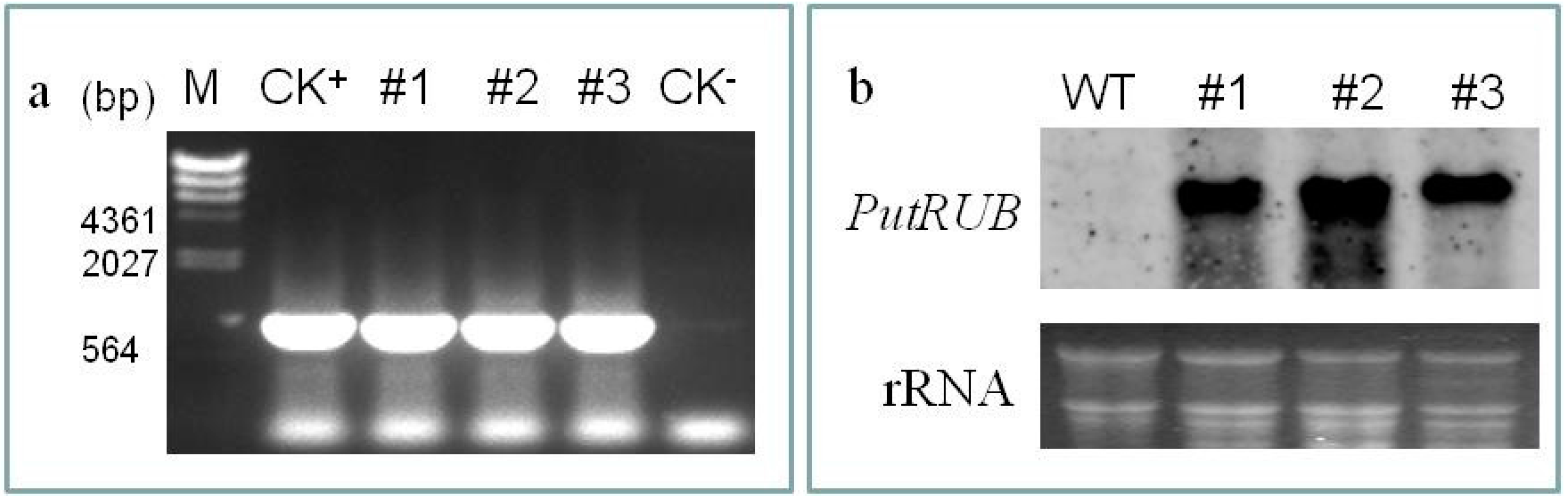
2.4. Response to NaCl and NaHCO3 Stress in PutRUB Transgenic Plants
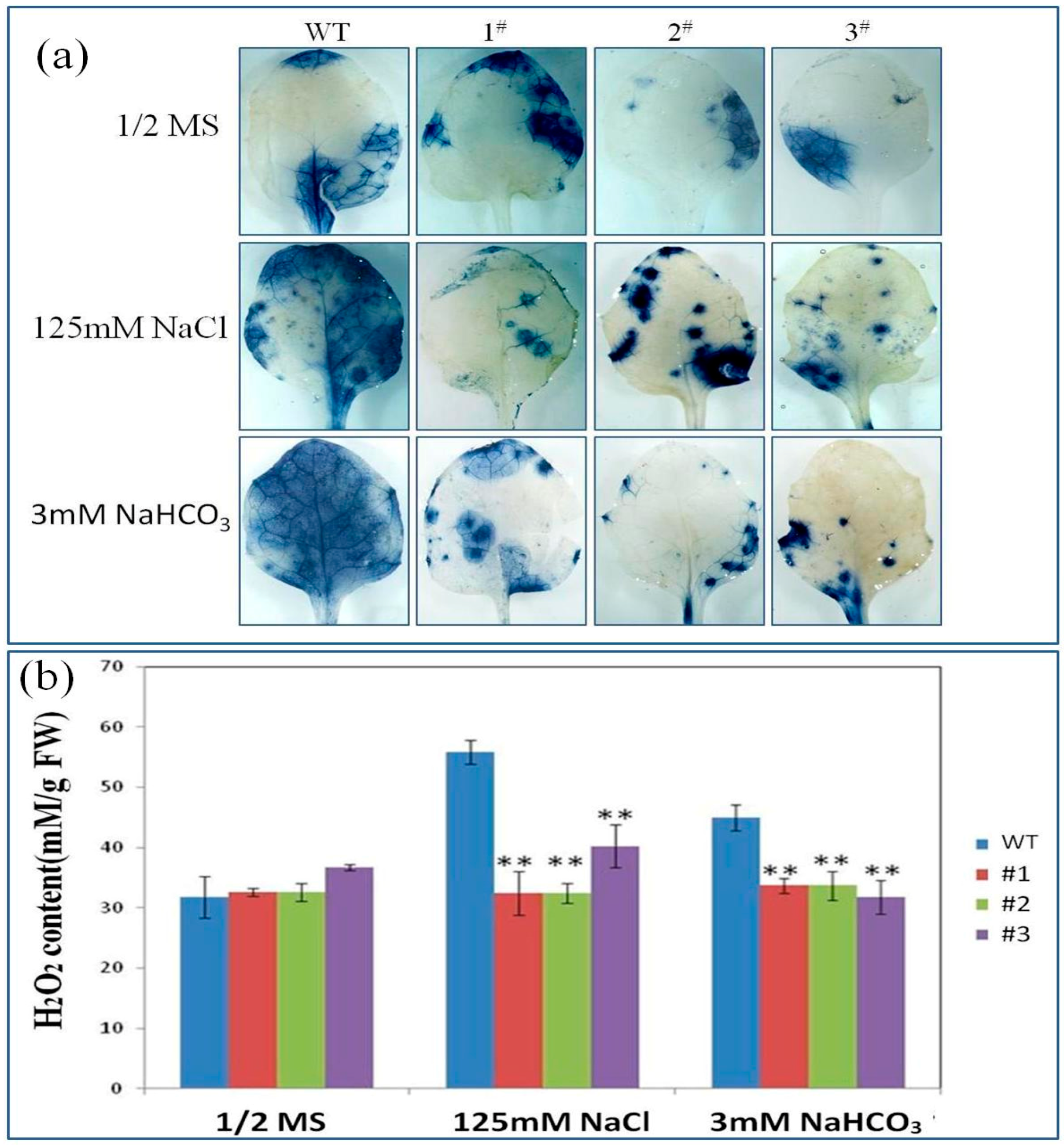
2.5. PutRUB Is Involved in Reducing of Reactive Oxygen Species (ROS) Detoxification
3. Materials and Methods
3.1. Plant Materials and Stress Treatments
3.2. Phylogenetic Analysis
3.3. Constructs
3.4. Subcellular Localization Assay
3.5. Quantitative Real-Time PCR
3.6. Stress Response Analysis in Transgenic Arabidopsis
3.7. ROS Detection
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef] [PubMed]
- Tuteja, N. Mechanisms of high salinity tolerance in plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [PubMed]
- Wang, L.; Seki, K.; Miyazaki, T.; Ishihama, Y. The causes of soil alkalinization in the songnen plain of northeast china. Paddy Water Environ. 2009, 7, 259–270. [Google Scholar] [CrossRef]
- Zhang, L.M.; Liu, X.G.; Qu, X.N.; Yu, Y.; Han, S.P.; Dou, Y.; Xu, Y.Y.; Jing, H.C.; Hao, D.Y. Early transcriptomic adaptation to Na2CO3 stress altered the expression of a quarter of the total genes in the maize genome and exhibited shared and distinctive profiles with nacl and high pH stresses. J. Integr. Plant Biol. 2013, 55, 1147–1165. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Cui, P.; Sun, H.; Guo, W.; Yang, C.; Jin, H.; Fang, B.; Shi, D. Comparative effects of salt and alkali stresses on organic acid accumulation and ionic balance of seabuckthorn (Hippophae rhamnoides L). Ind. Crops Prod. 2009, 30, 351–358. [Google Scholar] [CrossRef]
- Miller, G.; Suzuki, N.; Ciftci-Yilmaz, S.; Mittler, R. Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 2010, 33, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wei, L.; Wang, Z.; Wang, T. Physiological and molecular features of Puccinellia tenuiflora tolerating salt and alkaline-salt stress. J. Integr. Plant Biol. 2013, 55, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, S.; Zhao, Q.; Wang, T.; Yang, C.; Diaz, C.; Sun, G.; Dai, S. Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J. Proteome Res. 2011, 10, 3852–3870. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.Q.; Shi, D.C.; Wang, D.L. The key physiological response to alkali stress by the alkali-resistant halophyte Puccinellia tenuiflora is the accumulation of large quantities of organic acids and into the rhyzosphere. J. Agron. Crop Sci. 2010, 196, 123–135. [Google Scholar] [CrossRef]
- Guorong, S.; Yongzhen, P.; Hongbo, S.; Liye, C.; Xining, Z.; Haiyan, M.; Wenzhong, C.; Cunxu, W. Does puccinelia tenuiflora have the ability of salt exudation? Colloids Surf. B Biointerfaces 2005, 46, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.H.; Zhu, Y.F.; Mao, Y.Q.; Wang, S.M.; Su, W.A.; Tang, Z.C. Alkali grass resists salt stress through high [K+] and an endodermis barrier to Na+. J. Exp. Bot. 2004, 55, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.M.; Zhang, J.L.; Liu, X.S.; Li, Z.; Wu, G.Q.; Cai, J.Y.; Flowers, T.J.; Wang, S.M. Puccinellia tenuiflora maintains a low Na+ level under salinity by limiting unidirectional Na+ influx resulting in a high selectivity for K+ over Na+. Plant Cell Environ. 2009, 32, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, G.; Suo, B.; Chen, G.; Wang, J.; Yan, Y. Effects of Na2CO3 and NaCl stresses on the antioxidant enzymes of chloroplasts and chlorophyll fluorescence parameters of leaves of Puccinellia tenuiflora (Turcz.) scribn.et Merr. Acta Physiol. Plant. 2008, 30, 143–150. [Google Scholar] [CrossRef]
- Ardie, S.W.; Liu, S.; Takano, T. Expression of the AKT1-type K+ channel gene from Puccinellia tenuiflora, putakt1, enhances salt tolerance in Arabidopsis. Plant Cell Rep. 2010, 29, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Ardie, S.W.; Nishiuchi, S.; Liu, S.; Takano, T. Ectopic expression of the K+ channel β subunits from Puccinellia tenuiflora (KPutB1) and rice (KOB1) alters K+ homeostasis of yeast and Arabidopsis. Mol. Biotechnol. 2011, 48, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Ardie, S.W.; Xie, L.; Takahashi, R.; Liu, S.; Takano, T. Cloning of a high-affinity K+ transporter gene PutHKT2;1 from Puccinellia tenuiflora and its functional comparison with OsHKT2;1 from rice in yeast and Arabidopsis. J. Exp. Bot. 2009, 60, 3491–3502. [Google Scholar] [CrossRef] [PubMed]
- Chang-Qing, Z.; Shunsaku, N.; Shenkui, L.; Tetsuo, T. Characterization of two plasma membrane protein 3 genes (PutPMP3) from the alkali grass, Puccinellia tenuiflora, and functional comparison of the rice homologues, OsLti6a/b from rice. BMB Rep. 2008, 41, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, X.; Takano, T.; Liu, S. Characterization of a PutCAX1 gene from Puccinellia tenuiflora that confers Ca2+ and Ba2+ tolerance in yeast. Biochem. Biophys. Res. Commun. 2009, 383, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, R.; Wang, B.; Liu, G.; Yang, C.; Cheng, Y. Functional characterization of a plasma membrane Na+/H+ antiporter from alkali grass (Puccinellia tenuiflora). Mol. Biol. Rep. 2011, 38, 4813–4822. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Markley, J.L. NMR spectroscopic studies of paramagnetic proteins: Iron-sulfur proteins. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 209–237. [Google Scholar] [CrossRef] [PubMed]
- Lovenberg, W.; Sobel, B.E. Rubredoxin: A new electron transfer protein from clostridium pasteurianum. Proc. Natl. Acad. Sci. USA 1965, 54, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, P.J.A.; Coon, M.J. Enzymatic omega-oxidation. 3. Purification and properties of rubredoxin, a component of the omega-hydroxylation system of Pseudomonas oleovorans. J. Biol. Chem. 1968, 243, 329–334. [Google Scholar]
- Klenk, H.P.; Clayton, R.A.; Tomb, J.F.; White, O.; Nelson, K.E.; Ketchum, K.A.; Dodson, R.J.; Gwinn, M.; Hickey, E.K.; Peterson, J.D.; et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 1997, 390, 364–370. [Google Scholar] [PubMed]
- Das Gracas, D.A.; Ramos, R.T.J.; Araujo, A.C.V.; Zahlouth, R.; Carneiro, A.R.; Lopes, T.S.; Barauna, R.A.; Azevedo, V.; Schneider, M.P.C.; Pellizari, V.H.; et al. Complete genome of a methanosarcina mazei strain isolated from sediment samples from an amazonian flooded area. Genome Announc. 2013, 1, e00271-13. [Google Scholar] [CrossRef] [PubMed]
- Terpolilli, J.; Hill, Y.; Tian, R.; Howieson, J.; Brau, L.; Goodwin, L.; Han, J.; Liolios, K.; Huntemann, M.; Pati, A.; et al. Genome sequence of ensifer meliloti strain wsm1022; A highly effective microsymbiont of the model legume medicago truncatula a17. Stand. Genom. Sci. 2013, 9, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Eidsness, M.K.; Lin, I.J.; Gebel, E.B.; Youn, B.; Harley, J.L.; Machonkin, T.E.; Frederick, R.O.; Markley, J.L.; Smith, E.T.; et al. Crystallographic studies of v44 mutants of clostridium pasteurianum rubredoxin: Effects of side-chain size on reduction potential. Proteins Struct. Funct. Bioinform. 2004, 57, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Hagelueken, G.; Wiehlmann, L.; Adams, T.M.; Kolmar, H.; Heinz, D.W.; Tummler, B.; Schubert, W.D. Crystal structure of the electron transfer complex rubredoxin rubredoxin reductase of pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2007, 104, 12276–12281. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, M.Y.; LeGall, J.; Fareleira, P.; Santos, H.; Xavier, A.V. Rubredoxin oxidase, a new flavo-hemo-protein, is the site of oxygen reduction to water by the “strict anaerobe” desulfovibrio gigas. Biochem. Biophys. Res. Commun. 1993, 193, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Coulter, E.D.; Kurtz, D.M., Jr. A role for rubredoxin in oxidative stress protection in desulfovibrio vulgaris: Catalytic electron transfer to rubrerythrin and two-iron superoxide reductase. Arch. Biochem. Biophys. 2001, 394, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Jenney, F.E.; Verhagen, M.F.J.M.; Cui, X.; Adams, M.W.W. Anaerobic microbes: Oxygen detoxification without superoxide dismutase. Science 1999, 286, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Blake, P.R.; Park, J.B.; Bryant, F.O.; Aono, S.; Magnuson, J.K.; Eccleston, E.; Howard, J.B.; Summers, M.F.; Adams, M.W.W. Determinants of protein hyperthermostability: Purification and amino acid sequence of rubredoxin from the hyperthermophilic archaebacterium pyrococcus furiosus and secondary structure of the zinc adduct by NMR. Biochemistry 1991, 30, 10885–10895. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Takano, T.; Liu, S. Discovery and characterization of two novel salt-tolerance genes in Puccinellia tenuiflora. Int. J. Mol. Sci. 2014, 15, 16469–16483. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, S.; Wang, T.; Sun, G.; Dai, S. Comparative proteomic analysis of Puccinellia tenuiflora leaves under Na2CO3 stress. Int. J. Mol. Sci. 2013, 14, 1740–1762. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 2008, 13, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Fu, X.; Koo, Y.D.; Zhu, J.K.; Jenney, F.E., Jr.; Adams, M.W.; Zhu, Y.; Shi, H.; Yun, D.J.; Hasegawa, P.M.; et al. An enhancer mutant of Arabidopsis salt overly sensitive 3 mediates both ion homeostasis and the oxidative stress response. Mol. Cell. Biol. 2007, 27, 5214–5224. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Antonkine, M.L.; van der Est, A.; Vassiliev, I.R.; Brettel, K.; Bittl, R.; Zech, S.G.; Zhao, J.; Stehlik, D.; Bryant, D.A.; et al. Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of Fx in synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J. Biol. Chem. 2002, 277, 20355–20366. [Google Scholar] [CrossRef] [PubMed]
- Shen, G.; Zhao, J.; Reimer, S.K.; Antonkine, M.L.; Cai, Q.; Weiland, S.M.; Golbeck, J.H.; Bryant, D.A. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J. Biol. Chem. 2002, 277, 20343–20354. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Dat, J.; Vandenabeele, S.; Vranova, E.; Van Montagu, M.; Inze, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Oxidant and antioxidant signalling in plants: A re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005, 28, 1056–1071. [Google Scholar] [CrossRef]
- Gomez, J.M.; Hernandez, J.A.; Jimenez, A.; del Rio, L.A.; Sevilla, F. Differential response of antioxidative enzymes of chloroplasts and mitochondria to long-term nacl stress of pea plants. Free Radic. Res. 1999, 31, S11–S18. [Google Scholar] [CrossRef] [PubMed]
- Pawlak-Sprada, S.; Arasimowicz-Jelonek, M.; Podgorska, M.; Deckert, J. Activation of phenylpropanoid pathway in legume plants exposed to heavy metals. Part I. Effects of cadmium and lead on phenylalanine ammonia-lyase gene expression, enzyme activity and lignin content. Acta Biochim. Pol. 2011, 58, 211–216. [Google Scholar] [PubMed]
- Laloi, C.; Mestres-Ortega, D.; Marco, Y.; Meyer, Y.; Reichheld, J.P. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004, 134, 1006–1016. [Google Scholar] [CrossRef] [PubMed]
- Auchere, F.; Sikkink, R.; Cordas, C.; Raleiras, P.; Tavares, P.; Moura, I.; Moura, J.J. Overexpression and purification of treponema pallidum rubredoxin; kinetic evidence for a superoxide-mediated electron transfer with the superoxide reductase neelaredoxin. J. Biol. Inorg. Chem. 2004, 9, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’S conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef] [PubMed]
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins a2-bryson, vernon. In Evolving Genes and Proteins; Vogel, H.J., Ed.; Academic Press: San Jose, CA, USA, 1965; pp. 97–166. [Google Scholar]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D.W.; Sambrook, J. The Condensed Protocols from Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2006, 800p. [Google Scholar]
- Hoffmann, A.; Hammes, E.; Plieth, C.; Desel, C.; Sattelmacher, B.; Hansen, U.P. Effect of CO2 supply on formation of reactive oxygen species in Arabidopsis thaliana. Protoplasma 2005, 227, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Nag, S.; Saha, K.; Choudhuri, M.A. A rapid and sensitive assay method for measuring amine oxidase based on hydrogen peroxide-titanium complex formation. Plant Sci. Int. J. Exp. Plant Biol. 2000, 157, 157–163. [Google Scholar] [CrossRef]

| Primers Name | Sequences (5′→3′) |
|---|---|
| PutRUB-F | ATGGCTGCCACGCTCTCCTCTGT |
| PutRUB-R | CTATTGCAGGCCATATACAAGCAG |
| PutRUB-YE,PB-F | GGATCCATGGCTGCCACGCTCTCCTCTGT |
| PutRUB-YE,PB-R | CTCGAGCTATTGCAGGCCATATACAAGCAG |
| PutRUB-GFP-FW | GGTACCATGGCTGCCACGCTCTCCTCTGT |
| PutRUB-GFP-RV | GGATCCTTGCAGGCCATATACAAGCAG |
| PutRUB-QPCR-F | GTTCACCAAGACAGCCATCCAG |
| PutRUB-QPCR-R | CAGTCAGCTTCCGTCCAAATCG |
| Put-Tubulin-F | GTGTCAGCCATACTGTGCCAATC |
| Put-Tubulin-R | TTGCTCATGCGGTCAGCAATACC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Liu, P.; Takano, T.; Liu, S. A Chloroplast-Localized Rubredoxin Family Protein Gene from Puccinellia tenuiflora (PutRUB) Increases NaCl and NaHCO3 Tolerance by Decreasing H2O2 Accumulation. Int. J. Mol. Sci. 2016, 17, 804. https://doi.org/10.3390/ijms17060804
Li Y, Liu P, Takano T, Liu S. A Chloroplast-Localized Rubredoxin Family Protein Gene from Puccinellia tenuiflora (PutRUB) Increases NaCl and NaHCO3 Tolerance by Decreasing H2O2 Accumulation. International Journal of Molecular Sciences. 2016; 17(6):804. https://doi.org/10.3390/ijms17060804
Chicago/Turabian StyleLi, Ying, Panpan Liu, Tetsuo Takano, and Shenkui Liu. 2016. "A Chloroplast-Localized Rubredoxin Family Protein Gene from Puccinellia tenuiflora (PutRUB) Increases NaCl and NaHCO3 Tolerance by Decreasing H2O2 Accumulation" International Journal of Molecular Sciences 17, no. 6: 804. https://doi.org/10.3390/ijms17060804





