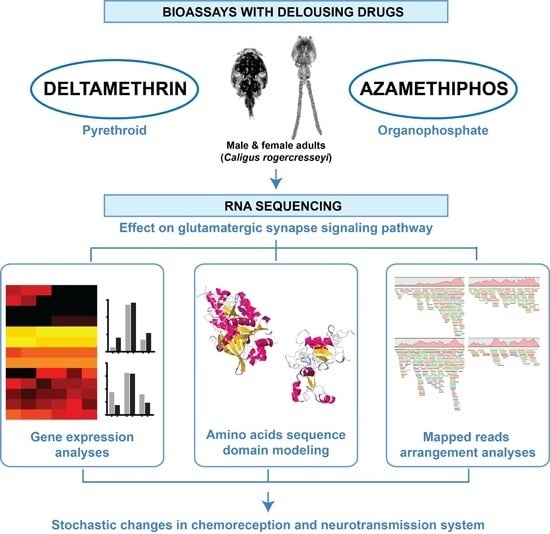
Pesticides Drive Stochastic Changes in the Chemoreception and Neurotransmission System of Marine Ectoparasites
Abstract
:1. Introduction
2. Results
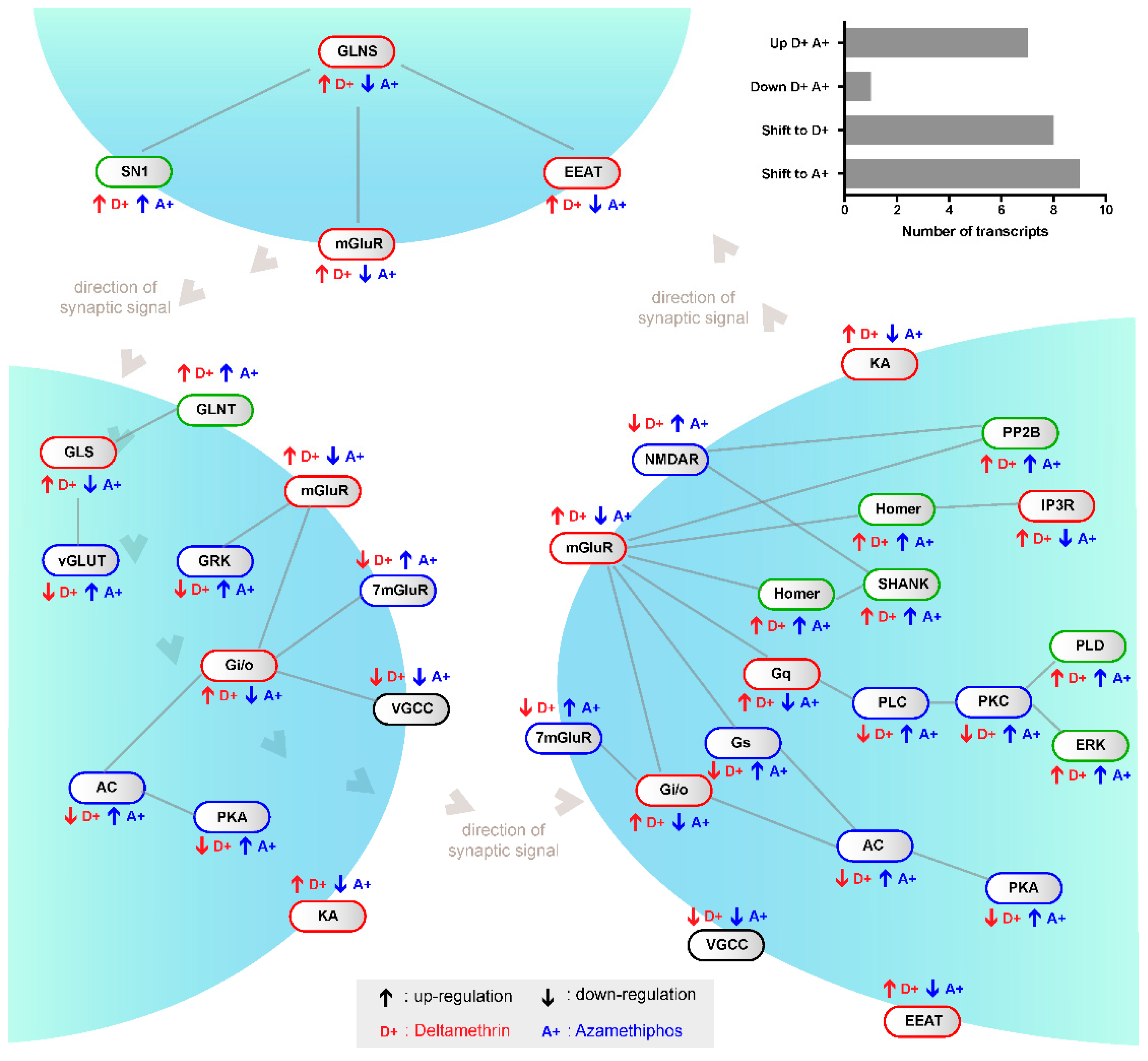
2.1. Transcriptomic Responses of Glutamatergic Pathway Components to Antiparasitic Treatment: A Global Comparison
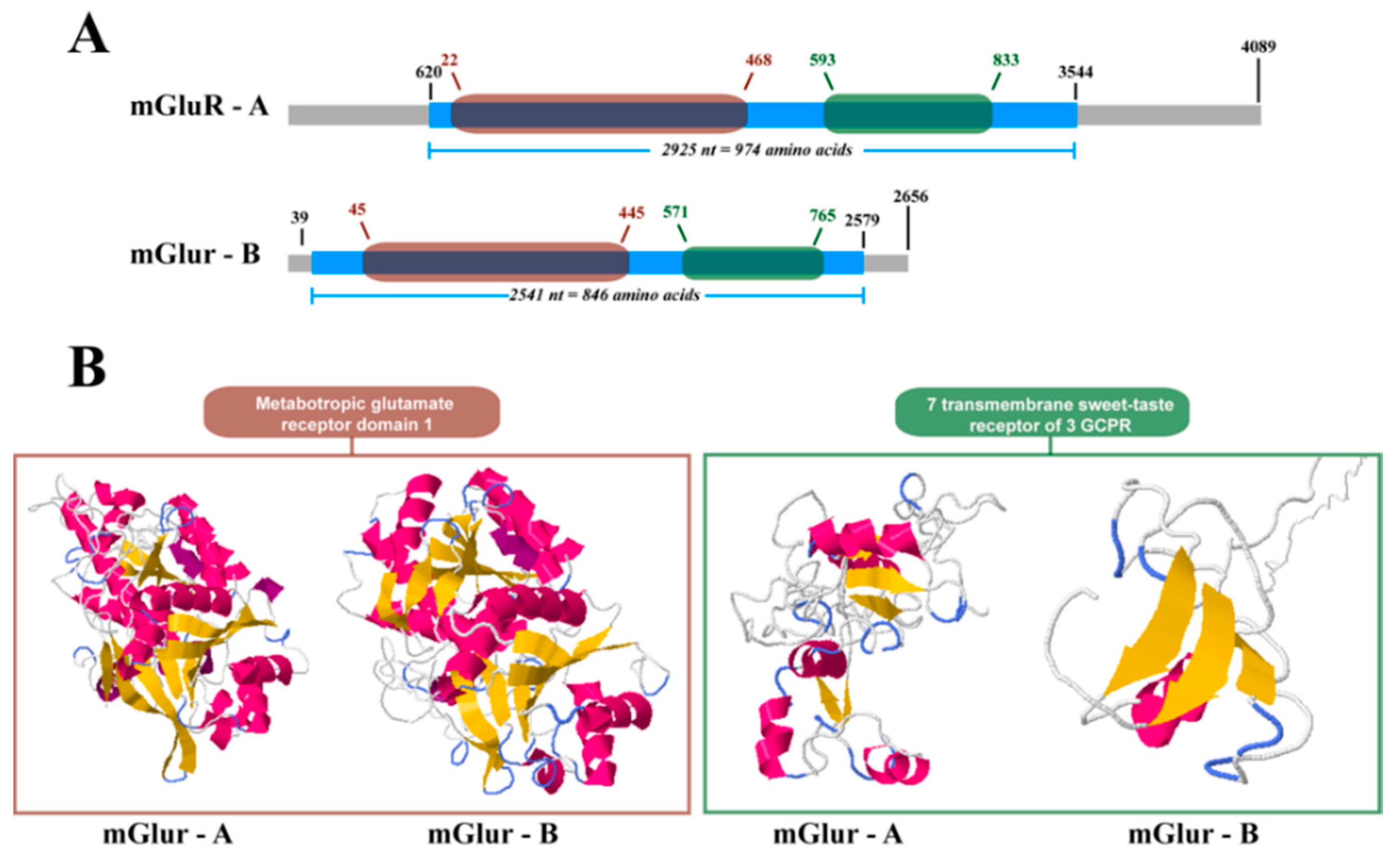
2.2. mGluR Characterization in Caligus Rogercresseyi
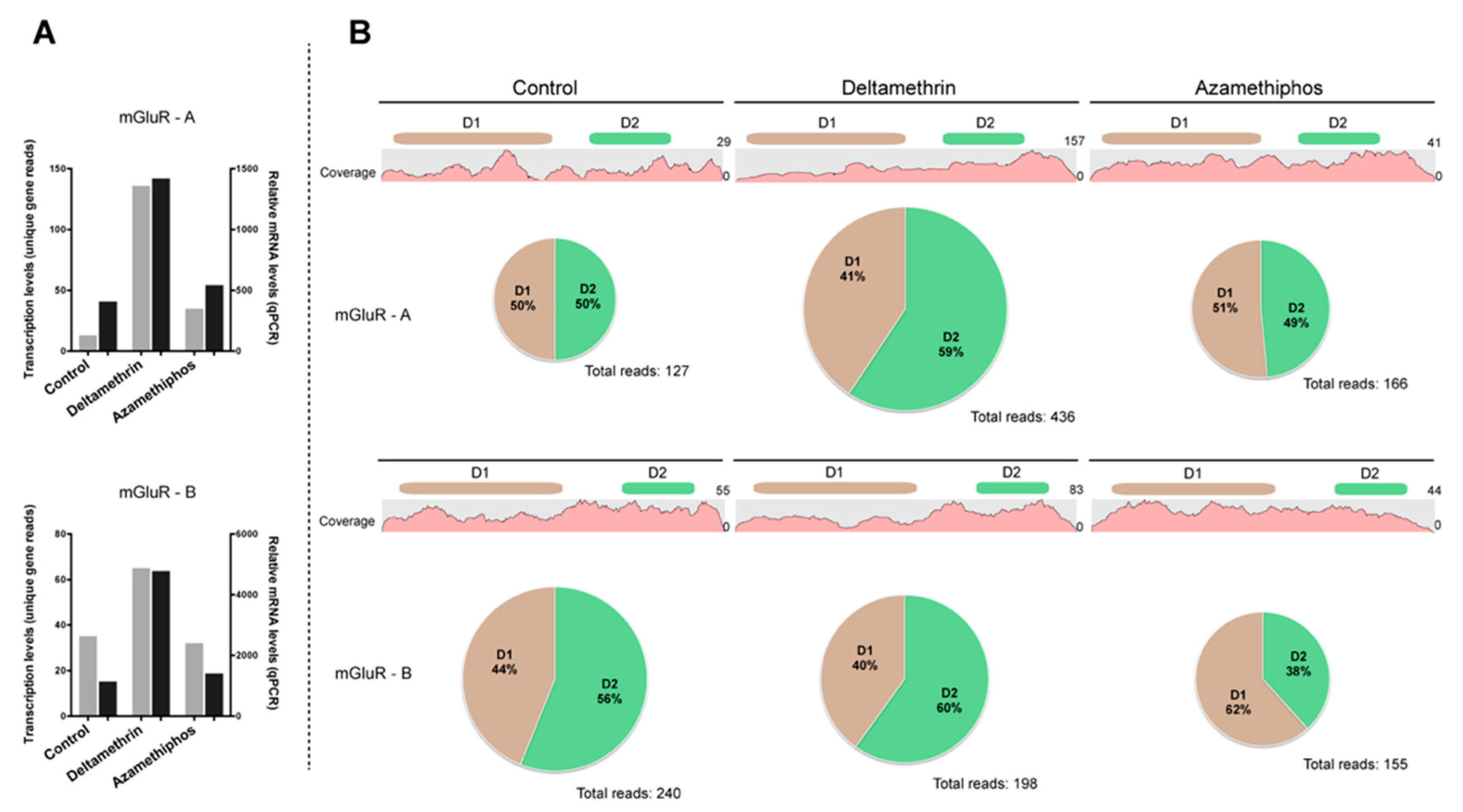
2.3. Gene Transcription of Cr-mGluR after Drug Treatments and Mapping Arrangement Analysis
3. Discussion
4. Materials and Methods
4.1. Bioassays and Transcriptome Sequencing
4.2. In Silico Analyses of Glutamatergic Synapse Pathway in Sea Lice to Antiparasitic Drugs
4.3. Statistical Analyses
4.4. Structural Analysis of Cr-mGluRs
4.5. Gene Transcription and Imbalance Analysis of mGluR
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DM | Deltamethrin |
| AZA | Azamethiphos |
| mRNA | messenger RNA |
| GPCRs | G protein-coupled receptors |
| mGluR | metabotropic receptor |
| iGluR | ionotropic glutamate receptor |
| IR | ionotropic receptor |
| RPKM | reads per kilobase of transcript per million mapped reads |
| SN1 | solute carrier family 8, member 3 |
| GLS | glutaminase (solute carrier family 8, member 2) |
| PP2B | protein phosphatase 2B |
| EEAT | solute carrier family 1, member 2 |
| KA | ionotropic kainate receptor |
| 7mGLuR | 7-transmembranse glutamate receptor domain containing protein |
| GLNS | glutamine synthetase/glutamate-ammonia ligase |
| PCA | principal component analysis |
| KEGG | Kyoto Encyclopedia of Genomes and Genes |
References
- Hay, M.E. Marine chemical ecology: Chemical signals and crues structure marine populations, communisties, and ecosystems. Annu. Rev. Mar. Sci. 2009, 1, 193–212. [Google Scholar] [CrossRef] [PubMed]
- Paul, V.J.; Ritson-Williams, R. Marine chemical ecology. Nat. Prod. Rep. 2008, 25, 662–695. [Google Scholar] [CrossRef] [PubMed]
- Rittschof, D.; Cohen, J.H. Crustacean peptide and peptide-like pheromones and kairomones. Peptides 2004, 25, 1503–1516. [Google Scholar] [CrossRef] [PubMed]
- Steiger, S.; Schmitt, T.; Scaefer, H. The origin and dynamic evolution of chemical information transfer. Proc. R. Soc. B 2011, 278, 970–979. [Google Scholar] [CrossRef] [PubMed]
- Burks, R.L.; Lodge, D.M. Cued in: Advances and opportunities in freshwater chemical ecology. J. Chem. Ecol. 2002, 28, 1901–1917. [Google Scholar] [CrossRef] [PubMed]
- Heuschele, J.; Selander, E. The chemical ecology of copepods. J. Plankton Res. 2014, 2014, 1–19. [Google Scholar] [CrossRef]
- Ingvarsdóttir, A.; Birkett, M.A.; Duce, I.; Mordue, W.; Pickett, J.A.; Wadhams, L.J.; Mordue, A.J. Role of semiochemicals in mate location by parasitic sea louse, lepeophtheirus salmonis. J. Chem. Ecol. 2002, 28, 2107–2117. [Google Scholar] [CrossRef] [PubMed]
- Bailey, R.J.E.; Birkett, M.A.; Ingvarsdóttir, A.; Mordue, A.J.; Mordue, W.; O’Shea, B.; Pickett, J.A.; Wadhams, L.J. The role of semiochemicals in host location and non-host avoidance by salmon louse (Lepeophtheirus salmonis) copepodids. Can. J. Fish. Aquat. Sci. 2006, 63, 448–456. [Google Scholar] [CrossRef]
- Mordue Luntz, A.J.; Birkett, M.A. A review of host finding behaviour in the parasitic sea louse, Lepeophtheirus salmonis (Caligidae: Copepoda). J. Fish. Dis. 2009, 32, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Pino-Marambio, J.; Mordue Luntz, A.J.; Birkett, M.; Carvajal, J.; Asencio, G.; Mellado, A.; Quiroz, A. Behavioural studies of host, non-host and mate location by the sea louse, caligus rogercresseyi boxshall & bravo, 2000 (copepoda: Caligidae). Aquaculture 2007, 271, 70–76. [Google Scholar]
- Huang, Y.; Thathiah, A. Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett. 2015, 589, 1607–1619. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, A.; Schioth, H.B. The role of G protein-coupled receptors in the early evolution of neurotransmission and the nervous system. J. Exp. Biol. 2015, 218, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Xiao, M.-Y.; Gustafsson, B.; Niu, Y.-P. Metabotropic glutamate receptors in the trafficking of ionotropic glutamate and GABAA receptors at central synapses. Curr. Neuropharmacol. 2006, 4, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Récasens, M.; Guiramand, J.; Aimar, R.; Abdulkarim, A.; Barbanel, G. Metabotropic glutamate receptors as drug targets. Curr. Drug Targets 2007, 8, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Abuin, L.; Bargeton, B.; Ulbrich, M.H.; Isacoff, E.Y.; Kellenberger, S.; Benton, R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 2011, 69, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Touhara, K.; Vosshall, L.B. Sensing odorants and pheromones with chemosensory receptors. Annu Rev. Physiol. 2009, 71, 307–332. [Google Scholar] [CrossRef] [PubMed]
- Corey, E.; Bobkov, Y.; Ukhanov, K.; Ache, B.W. Ionotropic crustacean olfactory receptors. PLoS ONE 2013, 8, e60551. [Google Scholar] [CrossRef] [PubMed]
- Stepanyan, R.; Day, K.; Urban, J.; Hardin, D.L.; Shetty, R.S.; Derby, C.D.; Ache, B.W.; Mcclintock, T.S.; Hardin, D.H. Gene expression and specificity in the mature zone of the lobster olfactory organ. Physiol. Genom. 2006, 25, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Núñez-Acuña, G.; Valenzuela-Muñoz, V.; Marambio, J.P.; Wadsworth, S.; Gallardo-Escárate, C. Insights into the olfactory system of the ectoparasite caligus rogercresseyi: Molecular characterization and gene transcription analysis of novel ionotropic receptors. Exp. Parasitol. 2014, 145, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Pomierny-Chamioło, L.; Rup, K.; Pomierny, B.; Niedzielska, E.; Kalivas, P.W.; Filip, M. Metabotropic glutamatergic receptors and their ligands in drug addiction. Pharmacol. Ther. 2014, 142, 281–305. [Google Scholar] [CrossRef] [PubMed]
- Salerno, G.; Colazza, S.; Conti, E. Sub-lethal effects of deltamethrin on walking behaviour and response to host kairomone of the egg parasitoid trissolcus basalis. Pest Manag. Sci. 2002, 58, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, X.; Gui, Q.-Q.; Xu, Q.-J. Sublethal effects of four insecticides on anagrus nilaparvatae (Hymenoptera: Mymaridae), an important egg parasitoid of the rice planthopper nilaparvata lugens (Homoptera: Delphacidae). Crop Protect. 2012, 37, 13–19. [Google Scholar] [CrossRef]
- Costello, M.J. The global economic cost of sea lice to the salmonid farming industry. J. Fish. Dis. 2009, 32, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Helgesen, K.; Bravo, S.; Sevatdal, S.; Mendoza, J.; Horsberg, T. Deltamethrin resistance in the sea louse caligus rogercresseyi (boxhall and bravo) in chile: Bioassay results and usage data for antiparasitic agents with references to norwegian conditions. J. Fish. Dis. 2014, 37, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Frank, T.; Friedrich, R.W. Neurobiology: Individuality sniffed out in flies. Nature 2015, 526, 200–201. [Google Scholar] [CrossRef] [PubMed]
- Laurent, G. Olfactory network dynamics and the coding of multidimensional signals. Nat. Rev. Neurosci. 2002, 3, 884–895. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Mardones, J.; Gallardo-Escárate, C. Next-generation transcriptome profiling of the salmon louse caligus rogercresseyi exposed to deltamethrin (alphamax™): Discovery of relevant genes and sex-related differences. Mar. Biotechnol. 2015, 17, 793–810. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Muñoz, V.; Chavez-Mardones, J.; Gallardo-Escárate, C. RNA-seq analysis evidences multiple gene responses in caligus rogercresseyi exposed to the anti-salmon lice drug azamethiphos. Aquaculture 2015, 446, 156–166. [Google Scholar] [CrossRef]
- Tang, Z.-Q.; Liu, Y.-W.; Shi, W.; Dinh, E.H.; Hamlet, W.R.; Curry, R.J.; Lu, Y. Activation of synaptic group ii metabotropic glutamate receptors induces long-term depression at gabaergic synapses in cns neurons. J. Neurosci. 2013, 33, 15964–15977. [Google Scholar] [CrossRef] [PubMed]
- Awad-Granko, H.; Jeffrey Conn, P. Activation of groups I or III metabotropic glutamate receptors inhibits excitatory transmission in the rat subthalamic nucleus. Neuropharmacology 2001, 41, 32–41. [Google Scholar] [CrossRef]
- Price, C.J.; Karayannis, T.; Pál, B.Z.; Capogna, M. Group II and III mGluRs-mediated presynaptic inhibition of epscs recorded from hippocampal interneurons of CA1 stratum lacunosum moleculare. Neuropharmacology 2005, 49, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela-Muñoz, V.; Sturm, A.; Gallardo-Escárate, C. Transcriptomic insights on the ABC transporter gene family in the salmon louse caligus rogercresseyi. Parasites Vectors 2015, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Mardones, J.; Boltaña, S.; Valenzuela-Muñoz, V.; Gallardo-Escárate, C. Evidence for the induction of key components of the notch signaling pathway via deltamethrin and azamethiphos treatment in the sea louse caligus rogercresseyi. Comp. Biochem. Phys. D 2015, in press. [Google Scholar]
- Kaur, K.; Helgesen, K.O.; Bakke, M.J.; Horsberg, T.E. Mechanism behind resistance against the organophosphate azamethiphos in salmon lice (Lepeophtheirus salmonis). PLoS ONE 2015, 10, e0124220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stäubert, C.; Bhuiyan, H.; Lindahl, A.; Broom, O.J.; Zhu, Y.; Islam, S.; Linnarsson, S.; Lehtiö, J.; Nordström, A. Rewired metabolism in drug-resistant leukemia cells: A metabolic switch hallmarked by reduced dependence on exogenous glutamine. J. Biol. Chem. 2015, 290, 8348–8359. [Google Scholar] [CrossRef] [PubMed]
- Suárez, I.; Bodega, G.; Fernández, B. Glutamine synthetase in brain: Effect of ammonia. Neurochem. Int. 2002, 41, 123–142. [Google Scholar] [CrossRef]
- Sinakevitch, I.; Grau, Y.; Strausfeld, N.; Birman, S. Dynamics of glutamatergic signaling in the mushroom body of young adult drosophila. Neural Dev. 2010, 5, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seyednasrollah, F.; Laiho, A.; Elo, L.L. Comparison of software packages for detecting differential expression in rna-seq studies. Brief. Bioinform. 2013, 16, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Pelechano, V.; Wei, W.; Steinmetz, L.M. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature 2013, 497, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.a.; Liu, X.S. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nature 2014, 15, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, E.L.; Jaszczyszyn, Y.; Thermes, C. Library preparation methods for next-generation sequencing: Tone down the bias. Exp. Cell Res. 2014, 322, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Marín, S.; Ibarra, R.; Medina, M.; Jansen, P. Sensitivity of Caligus rogercresseyi (boxshall and bravo 2000) to pyrethroids and azamethiphos measured using bioassay tests—A large scale spatial study. Prev. Vet. Med. 2015, in press. [Google Scholar]
- Bravo, S.; Treasurer, J.; Sepulveda, M.; Lagos, C. Effectiveness of hydrogen peroxide in the control of Caligus rogercresseyi in chile and implications for sea louse management. Aquaculture 2010, 303, 22–27. [Google Scholar] [CrossRef]
- Chavez-Mardones, J.; Gallardo-Escárate, C. Deltamethrin (alphamax™) reveals modulation of genes related to oxidative stress in the ectoparasite caligus rogercresseyi: Implications on delousing drug effectiveness. Aquaculture 2014, 433, 421–429. [Google Scholar] [CrossRef]
- Gallardo-Escárate, C.; Valenzuela-Muñoz, V.; Núñez-Acuña, G. RNA-seq analysis using de novo transcriptome assembly as a reference for the salmon louse caligus rogercresseyi. PLoS ONE 2014, 9, e92239. [Google Scholar]
- Boltaña, S.; Rey, S.; Roher, N.; Vargas, R.; Huerta, M.; Huntingford, F.A.; Goetz, F.W.; Moore, J.; Garcia-Valtanen, P.; Estepa, A.; et al. Behavioural fever is a synergic signal amplifying the innate immune response. Proc. R. Soc. B Biol. Sci. 2013, 280, 20131381. [Google Scholar] [CrossRef] [PubMed]
- Czerwinska, U. Dedal: Cytoscape 3.0 app for producing and morphing data-driven and structure-driven network layouts. BMC Syst. Biol. 2015, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
- Gorban, A.N.; Zinovyev, A. Principal graphs and manifolds. In Handbook of Research on Machine Learning Applications and Trends: Algorithms, Methods and Techniques; Olivas, E.S., Guererro, J.D.M., Sober, M.M., Benedito, J.R.M., López, A.J.S., Eds.; IGI Global: Valencia, Spain, 2009. [Google Scholar]
- Buchan, D.W.A.; Minneci, F.; Nugent, T.C.O.; Bryson, K.; Jones, D.T. Scalable web services for the psipred protein analysis workbench. Nucleic Acid Res. 2013, 41, W349–W357. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Núñez-Acuña, G.; Boltaña, S.; Gallardo-Escárate, C. Pesticides Drive Stochastic Changes in the Chemoreception and Neurotransmission System of Marine Ectoparasites. Int. J. Mol. Sci. 2016, 17, 700. https://doi.org/10.3390/ijms17060700
Núñez-Acuña G, Boltaña S, Gallardo-Escárate C. Pesticides Drive Stochastic Changes in the Chemoreception and Neurotransmission System of Marine Ectoparasites. International Journal of Molecular Sciences. 2016; 17(6):700. https://doi.org/10.3390/ijms17060700
Chicago/Turabian StyleNúñez-Acuña, Gustavo, Sebastián Boltaña, and Cristian Gallardo-Escárate. 2016. "Pesticides Drive Stochastic Changes in the Chemoreception and Neurotransmission System of Marine Ectoparasites" International Journal of Molecular Sciences 17, no. 6: 700. https://doi.org/10.3390/ijms17060700