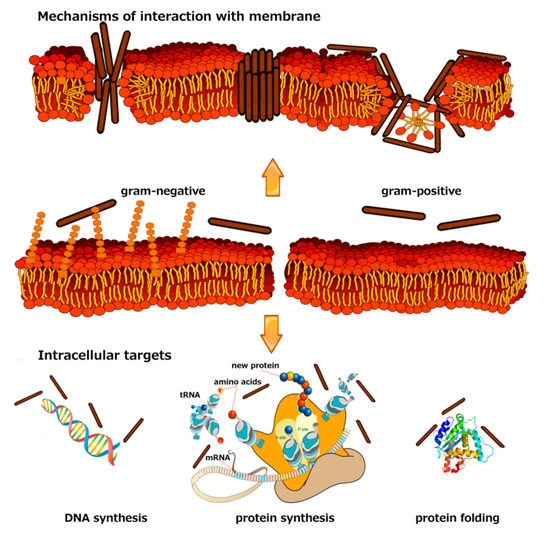
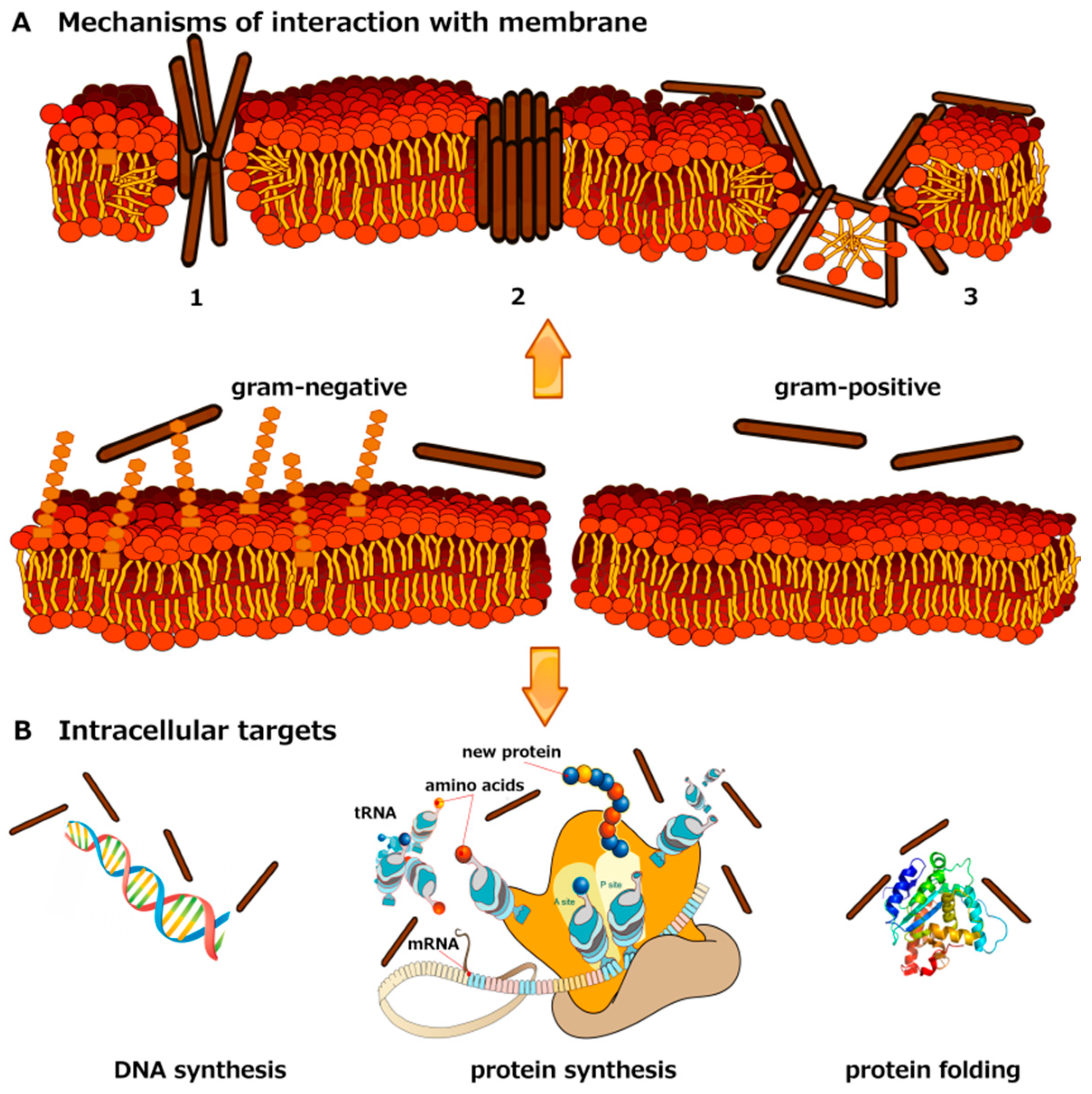
Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria
Abstract
:1. Introduction
2. Marine-Derived AMPs
3. Challenges in the Use of AMPs as Drugs
4. Chemical Modification of Marine AMPs
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Aiello, A.E.; Larson, E. Antibacterial cleaning and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect. Dis. 2003, 3, 501–506. [Google Scholar] [CrossRef]
- Arnold, S.R. Revenge of the killer microbe. CMAJ 2007, 177, 895–896. [Google Scholar] [CrossRef] [PubMed]
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Berisio, R.; Grieco, P.; Morelli, G.; Galdiero, M. Antimicrobial peptides as an opportunity against bacterial diseases. Curr. Med. Chem. 2015, 22, 1665–1677. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.; Ortiz, C.; Guzman, F.; Fernandez-Lafuente, R.; Torres, R. Antimicrobial peptides: Promising compounds against pathogenic microorganisms. Curr. Med. Chem. 2014, 21, 2299–2321. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Marr, A.K.; Gooderham, W.J.; Hancock, R.E. Antibacterial peptides for therapeutic use: Obstacles and realistic outlook. Curr. Opin. Pharmacol. 2006, 6, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. Amped up immunity: How antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Shai, Y. Mode of action of membrane active antimicrobial peptides. Pept. Sci. 2002, 66, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; de Vries, A.H.; Tieleman, D.P. Lipids on the move: Simulations of membrane pores, domains, stalks and curves. Biochim. Biophys. Acta 2009, 1788, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem. Biol. 2010, 5, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Falanga, A.; Galdiero, M.; Galdiero, S. Membranotropic cell penetrating peptides: The outstanding journey. Int. J. Mol. Sci. 2015, 16, 25323–25337. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Falanga, A.; Morelli, G.; Galdiero, M. Gh625: A milestone in understanding the many roles of membranotropic peptides. Biochim. Biophys. Acta 2015, 1848, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Vitiello, M.; Falanga, A.; Cantisani, M.; Incoronato, N.; Galdiero, M. Intracellular delivery: Exploiting viral membranotropic peptides. Curr. Drug Metab. 2012, 13, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Ponnappan, N.; Budagavi, D.P.; Yadav, B.K.; Chugh, A. Membrane-active peptides from marine organisms-antimicrobials, cell-penetrating peptides and peptide toxins: Applications and prospects. Probiotics Antimicrob. Proteins 2015, 7, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.K.; Seo, C.H.; Park, Y. Marine peptides and their anti-infective activities. Mar. Drugs 2015, 13, 618–654. [Google Scholar] [CrossRef] [PubMed]
- Charlet, M.; Chernysh, S.; Philippe, H.; Hetru, C.; Hoffmann, J.A.; Bulet, P. Innate immunity. Isolation of several cysteine-rich antimicrobial peptides from the blood of a mollusc, mytilus edulis. J. Biol. Chem. 1996, 271, 21808–21813. [Google Scholar] [PubMed]
- Cheung, R.C.; Ng, T.B.; Wong, J.H. Marine peptides: Bioactivities and applications. Mar. Drugs 2015, 13, 4006–4043. [Google Scholar] [CrossRef] [PubMed]
- Otero-González, A.J.; Magalhães, B.S.; Garcia-Villarino, M.; López-Abarrategui, C.; Sousa, D.A.; Dias, S.C.; Franco, O.L. Antimicrobial peptides from marine invertebrates as a new frontier for microbial infection control. FASEB J. 2010, 24, 1320–1334. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Lee, I.H.; Menzel, L.; Waring, A.; Zhao, C. Clavanins and styelins, α-helical antimicrobial peptides from the hemocytes of styela clava. Adv. Exp. Med. Biol. 2001, 484, 71–76. [Google Scholar] [PubMed]
- Pundir, P.; Catalli, A.; Leggiadro, C.; Douglas, S.E.; Kulka, M. Pleurocidin, a novel antimicrobial peptide, induces human mast cell activation through the fprl1 receptor. Mucosal Immunol. 2014, 7, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.C.; Lee, S.H.; Hour, A.-L.; Pan, C.-Y.; Lee, L.-H.; Chen, J.-Y. Five different piscidins from Nile Tilapia, Oreochromis niloticus: Analysis of their expressions and biological functions. PLoS ONE 2012, 7, e50263. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-A.; Kim, Y.K.; Lim, S.S.; Zhu, W.L.; Ko, H.; Shin, S.Y.; Hahm, K.-S.; Kim, Y. Solution structure and cell selectivity of piscidin 1 and its analogues. Biochemistry 2007, 46, 3653–3663. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.; Choi, H.; Lee, D.G. Influence of the N- and C-terminal regions of antimicrobial peptide pleurocidin on antibacterial activity. J. Microbiol. Biotechnol. 2012, 22, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.M.; Weis, P.; Diamond, G. Isolation and characterization of pleurocidin, an antimicrobial peptide in the skin secretions of winter flounder. J. Biol. Chem. 1997, 272, 12008–12013. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Y.; Chen, J.Y.; Cheng, Y.S.; Chen, C.Y.; Ni, I.H.; Sheen, J.F.; Pan, Y.L.; Kuo, C.M. Gene expression and localization of the epinecidin-1 antimicrobial peptide in the grouper (Epinephelus coioides), and its role in protecting fish against pathogenic infection. DNA Cell Biol. 2007, 26, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Cho, Y.; Lehrer, R.I. Styelins, broad-spectrum antimicrobial peptides from the solitary tunicate, styela clava. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1997, 118, 515–521. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Andrew Tincu, J.; Taylor, S.W.; Menzel, L.P.; Waring, A.J. Natural peptide antibiotics from tunicates: Structures, functions and potential uses. Integr. Comp. Biol. 2003, 43, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Galinier, R.; Roger, E.; Sautiere, P.E.; Aumelas, A.; Banaigs, B.; Mitta, G. Halocyntin and papillosin, two new antimicrobial peptides isolated from hemocytes of the solitary tunicate, Halocynthia papillosa. J. Pept. Sci. 2009, 15, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Lee, Y.S.; Kim, C.H.; Kim, C.R.; Hong, T.; Menzel, L.; Boo, L.M.; Pohl, J.; Sherman, M.A.; Waring, A.; et al. Dicynthaurin: An antimicrobial peptide from hemocytes of the solitary tunicate, Halocynthia aurantium. Biochim. Biophys. Acta 2001, 1527, 141–148. [Google Scholar] [CrossRef]
- Subramanian, S.; Ross, N.W.; MacKinnon, S.L. Myxinidin, a novel antimicrobial peptide from the epidermal mucus of hagfish, Myxine glutinosa L. Mar. Biotechnol. 2009, 11, 748–757. [Google Scholar] [CrossRef] [PubMed]
- Cantisani, M.; Finamore, E.; Mignogna, E.; Falanga, A.; Nicoletti, G.F.; Pedone, C.; Morelli, G.; Leone, M.; Galdiero, M.; Galdiero, S. Structural insights into and activity analysis of the antimicrobial peptide myxinidin. Antimicrob. Agents Chemother. 2014, 58, 5280–5290. [Google Scholar] [CrossRef] [PubMed]
- Cantisani, M.; Leone, M.; Mignogna, E.; Kampanaraki, K.; Falanga, A.; Morelli, G.; Galdiero, M.; Galdiero, S. Structure-activity relations of myxinidin, an antibacterial peptide derived from the epidermal mucus of hagfish. Antimicrob. Agents Chemother. 2013, 57, 5665–5673. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Park, C.B.; Kim, M.S.; Kim, S.C. Parasin i, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998, 437, 258–262. [Google Scholar] [CrossRef]
- Koo, Y.S.; Kim, J.M.; Park, I.Y.; Yu, B.J.; Jang, S.A.; Kim, K.S.; Park, C.B.; Cho, J.H.; Kim, S.C. Structure-activity relations of parasin I, a histone H2A-derived antimicrobial peptide. Peptides 2008, 29, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.H.; Cho, Y.; Lehrer, R.I. Effects of ph and salinity on the antimicrobial properties of clavanins. Infect. Immun. 1997, 65, 2898–2903. [Google Scholar] [PubMed]
- Van Kan, E.J.; van der Bent, A.; Demel, R.A.; de Kruijff, B. Membrane activity of the peptide antibiotic clavanin and the importance of its glycine residues. Biochemistry 2001, 40, 6398–6405. [Google Scholar] [CrossRef] [PubMed]
- Tasiemski, A.; Schikorski, D.; Le Marrec-Croq, F.; van Camp, C.P.; Boidin-Wichlacz, C.; Sautiere, P.E. Hedistin: A novel antimicrobial peptide containing bromotryptophan constitutively expressed in the NK cells-like of the marine annelid, Nereis diversicolor. Dev. Comp. Immunol. 2007, 31, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Stensvåg, K.; Haug, T.; Sperstad, S.V.; Rekdal, Ø.; Indrevoll, B.; Styrvold, O.B. Arasin 1, a proline–Arginine-rich antimicrobial peptide isolated from the spider crab, Hyas araneus. Dev. Comp. Immunol. 2008, 32, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, V.S.; Blencke, H.M.; Benincasa, M.; Haug, T.; Eksteen, J.J.; Styrvold, O.B.; Scocchi, M.; Stensvag, K. Structure-activity relationships of the antimicrobial peptide arasin 1—And mode of action studies of the N-terminal, proline-rich region. PLoS ONE 2013, 8, e53326. [Google Scholar] [CrossRef] [PubMed]
- Sperstad, S.V.; Haug, T.; Vasskog, T.; Stensvåg, K. Hyastatin, a glycine-rich multi-domain antimicrobial peptide isolated from the spider crab (Hyas araneus) hemocytes. Mol. Immunol. 2009, 46, 2604–2612. [Google Scholar] [CrossRef] [PubMed]
- Khoo, L.; Robinette, D.W.; Noga, E.J. Callinectin, an antibacterial peptide from blue crab, Callinectes sapidus, hemocytes. Mar. Biotechnol. 1999, 1, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Noga, E.J.; Stone, K.L.; Wood, A.; Gordon, W.L.; Robinette, D. Primary structure and cellular localization of callinectin, an antimicrobial peptide from the blue crab. Dev. Comp. Immunol. 2011, 35, 409–415. [Google Scholar] [CrossRef] [PubMed]
- Iijima, N.; Tanimoto, N.; Emoto, Y.; Morita, Y.; Uematsu, K.; Murakami, T.; Nakai, T. Purification and characterization of three isoforms of chrysophsin, a novel antimicrobial peptide in the gills of the red sea bream, Chrysophrys major. Eur. J. Biochem. 2003, 270, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tao, R.; Tong, Z.; Ding, Y.; Kuang, R.; Zhai, S.; Liu, J.; Ni, L. Effect of a novel antimicrobial peptide chrysophsin-1 on oral pathogens and streptococcus mutans biofilms. Peptides 2012, 33, 212–219. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.J.; Bertani, P.; Moulay, G.; Marquette, A.; Perrone, B.; Drake, A.F.; Kichler, A.; Bechinger, B. Membrane interaction of chrysophsin-1, a histidine-rich antimicrobial peptide from red sea bream. Biochemistry 2007, 46, 15175–15187. [Google Scholar] [CrossRef] [PubMed]
- Jiravanichpaisal, P.; Lee, S.Y.; Kim, Y.A.; Andren, T.; Soderhall, I. Antibacterial peptides in hemocytes and hematopoietic tissue from freshwater crayfish Pacifastacus leniusculus: Characterization and expression pattern. Dev. Comp. Immunol. 2007, 31, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Lee, B.L.; Soderhall, K. Processing of an antibacterial peptide from hemocyanin of the freshwater crayfish Pacifastacus leniusculus. J. Biol. Chem. 2003, 278, 7927–7933. [Google Scholar] [CrossRef] [PubMed]
- Ennaas, N.; Hammami, R.; Gomaa, A.; Bedard, F.; Biron, E.; Subirade, M.; Beaulieu, L.; Fliss, I. Collagencin, an antibacterial peptide from fish collagen: Activity, structure and interaction dynamics with membrane. Biochem. Biophys. Res. Commun. 2016, 473, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Scudiero, O.; Galdiero, S.; Cantisani, M.; Di Noto, R.; Vitiello, M.; Galdiero, M.; Naclerio, G.; Cassiman, J.J.; Pedone, C.; Castaldo, G.; et al. Novel synthetic, salt-resistant analogs of human β-defensins 1 and 3 endowed with enhanced antimicrobial activity. Antimicrob. Agents Chemother. 2010, 54, 2312–2322. [Google Scholar] [CrossRef] [PubMed]
- Scudiero, O.; Galdiero, S.; Nigro, E.; Del Vecchio, L.; Di Noto, R.; Cantisani, M.; Colavita, I.; Galdiero, M.; Cassiman, J.J.; Daniele, A.; et al. Chimeric β-defensin analogs, including the novel 3NI analog, display salt-resistant antimicrobial activity and lack toxicity in human epithelial cell lines. Antimicrob. Agents Chemother. 2013, 57, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Carmona, M.; Hubert, P.; Delvenne, P.; Herfs, M. Defensins: “Simple” antimicrobial peptides or broad-spectrum molecules? Cytokine Growth Factor Rev. 2015, 26, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Ruangsri, J.; Kitani, Y.; Kiron, V.; Lokesh, J.; Brinchmann, M.F.; Karlsen, B.O.; Fernandes, J.M. A novel β-defensin antimicrobial peptide in atlantic cod with stimulatory effect on phagocytic activity. PLoS ONE 2013, 8, e62302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.G.; Zhou, L.; Jin, J.Y.; Zhao, Z.; Lan, J.; Zhang, Y.B.; Zhang, Q.Y.; Gui, J.F. Antimicrobial activity-specific to Gram-negative bacteria and immune modulation-mediated NF-κB and SP1 of a medaka β-defensin. Dev. Comp. Immunol. 2009, 33, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Adhya, M.; Jeung, H.D.; Kang, H.-S.; Choi, K.-S.; Lee, D.S.; Cho, M. Cloning and localization of MCDEF, a defensin from manila clams (Ruditapes philippinarum). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2012, 161, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Balandin, S.V.; Aleshina, G.M.; Tagaev, A.A.; Leonova, Y.F.; Krasnodembsky, E.D.; Men‘shenin, A.V.; Kokryakov, V.N. Aurelin, a novel antimicrobial peptide from jellyfish Aurelia aurita with structural features of defensins and channel-blocking toxins. Biochem. Biophys. Res. Commun. 2006, 348, 514–523. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Panteleev, P.V.; Balandin, S.V.; Gizatullina, A.K.; Altukhov, D.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S.; Ovchinnikova, T.V. Recombinant expression and solution structure of antimicrobial peptide aurelin from jellyfish Aurelia aurita. Biochem. Biophys. Res. Commun. 2012, 429, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Kang, D.I.; Zhu, W.L.; Shin, S.Y.; Hahm, K.S.; Kim, Y. Solution structures and biological functions of the antimicrobial peptide, arenicin-1, and its linear derivative. Biopolymers 2007, 88, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Aleshina, G.M.; Balandin, S.V.; Krasnosdembskaya, A.D.; Markelov, M.L.; Frolova, E.I.; Leonova, Y.F.; Tagaev, A.A.; Krasnodembsky, E.G.; Kokryakov, V.N. Purification and primary structure of two isoforms of arenicin, a novel antimicrobial peptide from marine polychaeta Arenicola marina. FEBS Lett. 2004, 577, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Ovchinnikova, T.V.; Shenkarev, Z.O.; Nadezhdin, K.D.; Balandin, S.V.; Zhmak, M.N.; Kudelina, I.A.; Finkina, E.I.; Kokryakov, V.N.; Arseniev, A.S. Recombinant expression, synthesis, purification, and solution structure of arenicin. Biochem. Biophys. Res. Commun. 2007, 360, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Roch, P.; Yang, Y.; Toubiana, M.; Aumelas, A. NMR structure of mussel mytilin, and antiviral–antibacterial activities of derived synthetic peptides. Dev. Comp. Immunol. 2008, 32, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Mitta, G.; Hubert, F.; Noel, T.; Roch, P. Myticin, a novel cysteine-rich antimicrobial peptide isolated from haemocytes and plasma of the mussel Mytilus galloprovincialis. Eur. J. Biochem. 1999, 265, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Shike, H.; Lauth, X.; Westerman, M.E.; Ostland, V.E.; Carlberg, J.M.; Van Olst, J.C.; Shimizu, C.; Bulet, P.; Burns, J.C. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 2002, 269, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- Vidal-Dupiol, J.; Ladriere, O.; Destoumieux-Garzon, D.; Sautiere, P.E.; Meistertzheim, A.L.; Tambutte, E.; Tambutte, S.; Duval, D.; Foure, L.; Adjeroud, M.; et al. Innate immune responses of a scleractinian coral to vibriosis. J. Biol. Chem. 2011, 286, 22688–22698. [Google Scholar] [CrossRef] [PubMed]
- Kawano, K.; Yoneya, T.; Miyata, T.; Yoshikawa, K.; Tokunaga, F.; Terada, Y.; Iwanaga, S. Antimicrobial peptide, tachyplesin I, isolated from hemocytes of the horseshoe crab (Tachypleus tridentatus). NMR determination of the β-sheet structure. J. Biol. Chem. 1990, 265, 15365–15367. [Google Scholar] [PubMed]
- Nakamura, T.; Furunaka, H.; Miyata, T.; Tokunaga, F.; Muta, T.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 1988, 263, 16709–16713. [Google Scholar] [PubMed]
- Miyata, T.; Tokunaga, F.; Yoneya, T.; Yoshikawa, K.; Iwanaga, S.; Niwa, M.; Takao, T.; Shimonishi, Y. Antimicrobial peptides, isolated from horseshoe crab hemocytes, tachyplesin II, and polyphemusins I and II: Chemical structures and biological activity. J. Biochem. 1989, 106, 663–668. [Google Scholar] [PubMed]
- Destoumieux, D.; Munoz, M.; Bulet, P.; Bachere, E. Penaeidins, a family of antimicrobial peptides from penaeid shrimp (Crustacea, Decapoda). Cell. Mol. Life Sci. 2000, 57, 1260–1271. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Haug, T.; Styrvold, O.B.; Jorgensen, T.O.; Stensvag, K. Strongylocins, novel antimicrobial peptides from the green sea urchin, strongylocentrotus droebachiensis. Dev. Comp. Immunol. 2008, 32, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Relf, J.M.; Chisholm, J.R.; Kemp, G.D.; Smith, V.J. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1999, 264, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Imjongjirak, C.; Amparyup, P.; Tassanakajon, A.; Sittipraneed, S. Molecular cloning and characterization of crustin from mud crab Scylla paramamosain. Mol. Biol. Rep. 2009, 36, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Song, C.; Liu, Y.; Wang, S.; Li, Q.; Li, X. Crustins from eyestalk cdna library of swimming crab Portunus trituberculatus: Molecular characterization, genomic organization and expression analysis. Fish Shellfish Immunol. 2012, 33, 937–945. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.Q.; Shi, Y.H.; Wang, Q. Characterisation of a novel type i crustin involved in antibacterial and antifungal responses in the red claw crayfish, Cherax quadricarinatus. Fish Shellfish Immunol. 2016, 48, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Du, X.J.; Xu, W.T.; Zhang, H.W.; Zhao, X.F.; Wang, J.X. Molecular cloning and characterization of three crustins from the chinese white shrimp, Fenneropenaeus chinensis. Fish Shellfish Immunol. 2010, 28, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Fernandes, J.M.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic wap domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [PubMed]
- Rolland, J.L.; Abdelouahab, M.; Dupont, J.; Lefevre, F.; Bachère, E.; Romestand, B. Stylicins, a new family of antimicrobial peptides from the pacific blue shrimp Litopenaeus stylirostris. Mol. Immunol. 2010, 47, 1269–1277. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.T.; Wang, J.; Mao, Y.; Liu, M.; Niu, S.F.; Qiao, Y.; Su, Y.Q.; Wang, C.Z.; Zheng, Z.P. Identification and expression analysis of a novel stylicin antimicrobial peptide from kuruma shrimp (Marsupenaeus japonicus). Fish Shellfish Immunol. 2015, 47, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, S.; Fusetani, N.; Konosu, S. Bioactive marine metabolites, IV. Isolation and the amino acid composition of discodermin a, an antimicrobial peptide, from the marine sponge Discodermia kiiensis. J. Nat. Prod. 1985, 48, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Houston, M.E., Jr.; Kondejewski, L.H.; Karunaratne, D.N.; Gough, M.; Fidai, S.; Hodges, R.S.; Hancock, R.E. Influence of preformed α-helix and α-helix induction on the activity of cationic antimicrobial peptides. J. Pept. Res. 1998, 52, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Avan, I.; Hall, C.D.; Katritzky, A.R. Peptidomimetics via modifications of amino acids and peptide bonds. Chem. Soc. Rev. 2014, 43, 3575–3594. [Google Scholar] [CrossRef] [PubMed]
- Danial, M.; van Dulmen, T.H.; Aleksandrowicz, J.; Potgens, A.J.; Klok, H.A. Site-specific pegylation of HR2 peptides: Effects of PEG conjugation position and chain length on HIV-1 membrane fusion inhibition and proteolytic degradation. Bioconjug. Chem. 2012, 23, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- Papo, N.; Oren, Z.; Pag, U.; Sahl, H.G.; Shai, Y. The consequence of sequence alteration of an amphipathic α-helical antimicrobial peptide and its diastereomers. J. Biol. Chem. 2002, 277, 33913–33921. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Carvalho, I.F.; Montelaro, R.C.; Gomes, P.; Martins, M.C. Covalent immobilization of antimicrobial peptides (AMPS) onto biomaterial surfaces. Acta Biomater. 2011, 7, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Post-translational modifications of natural antimicrobial peptides and strategies for peptide engineering. Curr. Biotechnol. 2012, 1, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Shinnar, A.E.; Butler, K.L.; Park, H.J. Cathelicidin family of antimicrobial peptides: Proteolytic processing and protease resistance. Bioorgan. Chem. 2003, 31, 425–436. [Google Scholar] [CrossRef]
- Yang, Y.S.; Mitta, G.; Chavanieu, A.; Calas, B.; Sanchez, J.F.; Roch, P.; Aumelas, A. Solution structure and activity of the synthetic four-disulfide bond mediterranean mussel defensin (MGD-1). Biochemistry 2000, 39, 14436–14447. [Google Scholar] [CrossRef] [PubMed]
- Noga, E.J.; Silphaduang, U.; Park, N.G.; Seo, J.K.; Stephenson, J.; Kozlowicz, S. Piscidin 4, a novel member of the piscidin family of antimicrobial peptides. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2009, 152, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.W.; Craig, A.G.; Fischer, W.H.; Park, M.; Lehrer, R.I. Styelin D, an extensively modified antimicrobial peptide from ascidian hemocytes. J. Biol. Chem. 2000, 275, 38417–38426. [Google Scholar] [CrossRef] [PubMed]
- Fedders, H.; Michalek, M.; Grotzinger, J.; Leippe, M. An exceptional salt-tolerant antimicrobial peptide derived from a novel gene family of haemocytes of the marine invertebrate Ciona intestinalis. Biochem. J. 2008, 416, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Goldman, M.J.; Anderson, G.M.; Stolzenberg, E.D.; Kari, U.P.; Zasloff, M.; Wilson, J.M. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell 1997, 88, 553–560. [Google Scholar] [CrossRef]
- Scudiero, O.; Nigro, E.; Cantisani, M.; Colavita, I.; Leone, M.; Mercurio, F.A.; Galdiero, M.; Pessi, A.; Daniele, A.; Salvatore, F.; et al. Design and activity of a cyclic mini-β-defensin analog: A novel antimicrobial tool. Int. J. Nanomed. 2015, 10, 6523–6539. [Google Scholar]
- Mai, J.; Tian, X.-L.; Gallant, J.W.; Merkley, N.; Biswas, Z.; Syvitski, R.; Douglas, S.E.; Ling, J.; Li, Y.-H. A novel target-specific, salt-resistant antimicrobial peptide against the cariogenic pathogen streptococcus mutans. Antimicrob. Agents Chem. 2011, 55, 5205–5213. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.H.; Ribeiro, S.M.; Nolasco, D.O.; de la Fuente-Nunez, C.; Felicio, M.R.; Goncalves, S.; Matos, C.O.; Liao, L.M.; Santos, N.C.; Hancock, R.E.; et al. A polyalanine peptide derived from polar fish with anti-infectious activities. Sci. Rep. 2016, 6, 21385. [Google Scholar] [CrossRef] [PubMed]
- Cantisani, M.; Vitiello, M.; Falanga, A.; Finamore, E.; Galdiero, M.; Galdiero, S. Peptides complementary to the active loop of porin P2 from haemophilus influenzae modulate its activity. Int. J. Nanomed. 2012, 7, 2361–2371. [Google Scholar]
- Franci, G.; Falanga, A.; Galdiero, S.; Palomba, L.; Rai, M.; Morelli, G.; Galdiero, M. Silver nanoparticles as potential antibacterial agents. Molecules 2015, 20, 8856–8874. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, S.; Capasso, D.; Vitiello, M.; D’Isanto, M.; Pedone, C.; Galdiero, M. Role of surface-exposed loops of haemophilus influenzae protein P2 in the mitogen-activated protein kinase cascade. Infect. Immun. 2003, 71, 2798–2809. [Google Scholar] [CrossRef] [PubMed]
- Notomista, E.; Falanga, A.; Fusco, S.; Pirone, L.; Zanfardino, A.; Galdiero, S.; Varcamonti, M.; Pedone, E.; Contursi, P. The identification of a novel sulfolobus islandicus CAMP-like peptide points to archaeal microorganisms as cell factories for the production of antimicrobial molecules. Microb. Cell Factories 2015, 14, 126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Int. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Bradshaw, J. Cationic antimicrobial peptides: Issues for potential clinical use. BioDrugs 2003, 17, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.N.; Rajanbabu, V.; Pan, C.Y.; Chan, Y.L.; Wu, C.J.; Chen, J.Y. Use of the antimicrobial peptide epinecidin-1 to protect against MRSA infection in mice with skin injuries. Biomaterials 2013, 34, 10319–10327. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.Y.; Rajanbabu, V.; Chen, J.Y.; Her, G.M.; Nan, F.H. Evaluation of the epinecidin-1 peptide as an active ingredient in cleaning solutions against pathogens. Peptides 2010, 31, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, I.E.; Morrison, A.E.; Cobb, J.E.; Fahey, C.A.; Camesano, T.A. Creating antibacterial surfaces with the peptide chrysophsin-1. ACS Appl. Mater. Interfaces 2012, 4, 5891–5897. [Google Scholar] [CrossRef] [PubMed]
- Dusane, D.H.; Pawar, V.S.; Nancharaiah, Y.V.; Venugopalan, V.P.; Kumar, A.R.; Zinjarde, S.S. Anti-biofilm potential of a glycolipid surfactant produced by a tropical marine strain of serratia marcescens. Biofouling 2011, 27, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.P.; Martin, M.M.; Goosney, D.L.; Hancock, R.E. The antimicrobial peptide polyphemusin localizes to the cytoplasm of Escherichia coli following treatment. Antimicrob. Agents Chemother. 2006, 50, 1522–1524. [Google Scholar] [CrossRef] [PubMed]
| Structure | Example of Marine AMPs | Organism | Microorganisms: Bacteria | Antibacterial Activity | Length | Ref. |
|---|---|---|---|---|---|---|
 Linear α-helical | Clavanins A, B, C, D and E | Solitary tunicate: Styela clava | Gram-negative: E. coli, S. typhimurium, P. aeruginosa | 0.4–5 μM (MIC) | 23 | [40] |
| Gram-positive: S. aureus, M. flavus | ||||||
| Dicynthaurin | Solitary tunicate: Halocynthia aurantium | Gram-negative: E. coli, P. aeruginosa | 140 μg/mL (MIC) | 60 | [34] | |
| Gram-positive: M. luteus, L. monocytogenes, S. aureus | ||||||
| Halocyntin | Ascidian: Halocynthia papillosa | Gram-negative: E. coli, P. aeruginosa, S. typhimurium, K. pneumoniae | 0.75–100 μM (MBC) | 26 | [33] | |
| Gram-positive: M. luteus, B. megaterium, A. viridans, S. aureus, E. facecolis | ||||||
| Hedistin | Annelid: Nereis diversicolor | Gram-negative: V. alginolyticus | 0.4–1.6 μM (MIC) | 22 | [42] | |
| Gram positive: M. luteus, M. nishinomiyaensis, S. aureus | ||||||
| Myxinidin | Hagfish: Myxine glutinosa | Gram-negative: E. coli, P. aeruginosa, S. Typhimurium, K. Pneumoniae | 5–30 μM (MIC) | 12 | [35,36,37] | |
| Gram-positive: S. aureus | ||||||
| Papillosin | Ascidian: Halocynthia papillosa | Gram-negative: E. coli | 0.25–1 μM (MBC) | 34 | [33] | |
| Gram-positive: M. luteus, S. aureus | ||||||
| Parasin 1 | Catfish: Parasilurus asotus | Gram-negative: E. coli, P. putida, S. enteriotidis | 2–4 μg/mL (MIC) | 19 | [38,39] | |
| Gram-positive: B. subtilis, S. aureus, S. mutans | ||||||
| Pleurocidin | Winter flounder: Pleuronectes americanus | Gram-negative: E. coli, C. aquatilis, S. typhimurium | 1.1–17.7 μM (MIC) | 25 | [28,29] | |
| Gram-positve: B. subtilis, S. aureus | ||||||
| Piscidins 1, 2, 3, 4 and 5 | FISH: Nila tilapia | Gram-negative: E. coli, P. vulgaris, V. vulnificus, P. aeruginosa | 0.6–23.6 μg/mL (MIC) | 21–44 | [26] | |
| Gram-positive: S. aureus, B. cereus, E. faecalis | ||||||
| Styelins A, B, C, D and E | Solitary tunicate: Styela clava | Gram-negative: E. coli, S. typhimurium, P. aeruginosa | 1–3 μg/mL (MIC) | 32 | [31,32] | |
| Gram-positive: L. monocytogenes, E. faecium, S. aureus | ||||||
 Linear or helical with abundance of one amino acid | Arasin 1 (proline and arginine rich) | Spider crab: Hyas araneus | Gram-negative: L. anguillarum, E. coli | 0.8–12.5 μM (MIC) | 37 | [43,44] |
| Gram-positive: C. glutamicum | ||||||
| Astacidin 1 and 2 (proline and arginine rich) | Crayfish: P. leniusculus | Gram-negative: S. flexneri, P. vulgaris, E. coli, P. aeruginosa | 0.5–15 μM (MIC) | 14–16 | [51,52] | |
| Gram-positive: S. aureus, B. megaterium, B. subtilis, M. luteus | ||||||
| Callinectin (proline, arginine and cysteine rich) | Blue crab: Callinectes sapidus | Gram-negative: E. coli | 1.44 μM (MBC) | 32 | [46,47] | |
| Chrysophsin 1, 2 and 3 (histidine rich) | Red Sea Bream: Chrysophrys major | Gram-negative: E. coli | 4–16 μg/mL (MIC) | 20–25 | [48,49,50] | |
| Gram-positive: B. subtilis, S. mutans | ||||||
| Hyastatin (glycine rich) | Spider crab: Hyas araneus | Gram-negative: E. coli | 0.4–12.5 μM (MIC) | 131 | [45] | |
| Gram-positive: C. glutamicus | ||||||
 Forming hairpin-like β-sheet or α-helical-β-sheet mixed structures with disulfide bonds | Arenicins 1, 2 and 3 | Polychaete: Arenicola marina | Gram-negative: E. coli, P. aeruginosa | 2–8 μM (MIC) | 21 | [62,63,64] |
| Gram-positive: L. monocytogenes, S. aureus, S. epidermidis | ||||||
| Aurelin | Jelly fish: Aurelia aurita | Gram-negative: E. coli | 7.6–22.6 μg/mL (MIC) | 40 | [60,61] | |
| Gram-positive: L. monocytogenes | ||||||
| Crustins types I, II and III | Crab: Carcinus maenas; Portunus trituberculatus | Gram-negative: P. aeruginosa, V. alginolyticus | 1.5–49.6 μM (MIC) | 77–95 | [75,76] | |
| Gram-positive: M. luteus, S. aureus | ||||||
| Damicornin | Coral: Pocillopora damicornis | Gram-positive: M. luteus, B. megaterium, S. aureus, B. stationis, M. maritypicum | 1.25–20 μM (MIC) | 40 | [68] | |
| Hepcidins | Seabream: Sparus aurata | Gram-negative: E. coli, A. hydrophila, V. prahaemloyticus | 6–24 μM (MIC) | 26 | [67] | |
| Gram-positive: B. subtilis, M. luteus, S. aureus | ||||||
| MCdef | Manila clams: Ruditapes philippinarum | Gram-negative: V. logei, V. salmonicida | 1.25–20 μM (MIC) | 44 | [59] | |
| Gram-positive: S. aureus, S. iniae | ||||||
| Myticins A, B and C | Mussel: Mytilus galloprovincialis | Gram-negative: E. coli, S. typhimurium, P. aeruginosa | 2.25–20 μM (MBC) | 40 | [66] | |
| Gram-positive: M. luteus, B. megaterium, E. viridans | ||||||
| Mylitin | Mussel: Mytilus galloprovincialis | Gram-negative: V. splendidus, V. anguillarum, E. coli | 125 μM–2 mM (MIC) | 34 | [65] | |
| Gram-positive: M. lysodeikticus | ||||||
| Penaeidins 1, 2 and 3 | shrimp: Penaeus vannamei | Gram-positive: B. megaterium, A. viridans | 0.3–2.5 μM (MIC) | 50–60 | [72] | |
| Poliphemusin | American horseshoe crab: Limulus polyphemus | Gram negative: E. coli | 0.25 μM (MIC) | 18 | [71,108] | |
| Strongylocins 1 and 2 | green sea urchin: Strongylocentrotus droebachiensis | Gram-negative: E. coli, L. anguillarum | 1.3–5 μM (MIC) | 83–90 | [73] | |
| Gram-positive: S. aureus, C. glutamicum | ||||||
| Tachyplesins I, II and III | Horseshoe Crab: Tachypleus tridentatus | Gram-negative: E. coli, S. thyphimurium | 0.6–6.3 μg/mL (MIC) | 17–18 | [69,70,71] | |
| Gram-positive: S. aureus | ||||||
| Cyclic peptides | Discodermin A | Marine sponge: Discodermia kiiensis | Gram-negative: P. mirabilis, P. morganii | 1.56–12.5 μg/mL (MIC) | 14 | [82] |
| Gram-positve: B. subtilis |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falanga, A.; Lombardi, L.; Franci, G.; Vitiello, M.; Iovene, M.R.; Morelli, G.; Galdiero, M.; Galdiero, S. Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria. Int. J. Mol. Sci. 2016, 17, 785. https://doi.org/10.3390/ijms17050785
Falanga A, Lombardi L, Franci G, Vitiello M, Iovene MR, Morelli G, Galdiero M, Galdiero S. Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria. International Journal of Molecular Sciences. 2016; 17(5):785. https://doi.org/10.3390/ijms17050785
Chicago/Turabian StyleFalanga, Annarita, Lucia Lombardi, Gianluigi Franci, Mariateresa Vitiello, Maria Rosaria Iovene, Giancarlo Morelli, Massimiliano Galdiero, and Stefania Galdiero. 2016. "Marine Antimicrobial Peptides: Nature Provides Templates for the Design of Novel Compounds against Pathogenic Bacteria" International Journal of Molecular Sciences 17, no. 5: 785. https://doi.org/10.3390/ijms17050785